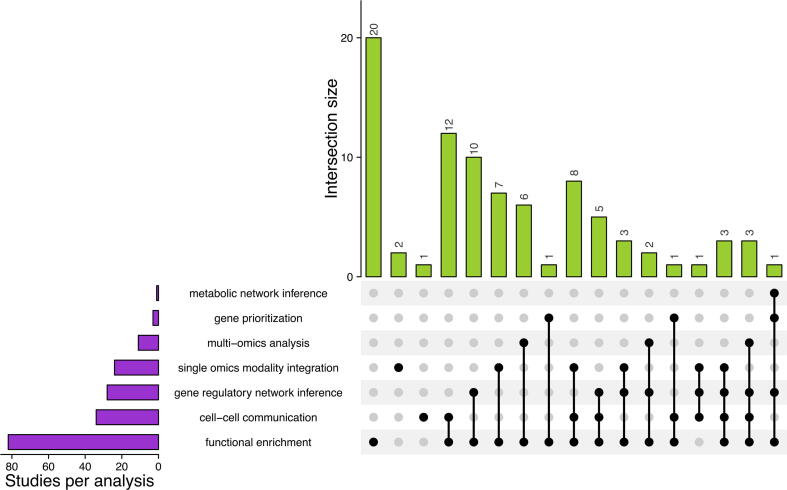
Fig. 1.
UpSet Plot showing the classification of studies characterizing single-cell EC heterogeneity with respect to the applied computational techniques. A total of 87 studies detailed in Supplementary Table 1, characterize single-cell EC heterogeneity with the distribution of studies that use different task-specific computational techniques. Performing differential expression of biomolecular abundances between conditions and subsequent coupling with functional enrichment techniques are commonly used to discover novel biological knowledge in single-cell ECs (82 studies). This is followed by the use of biological network inference techniques to identify novel biomolecular interactions from changes in gene expression (18 studies). Within biological network inference approaches, most studies intend to predict cell–cell communication through ligand-receptor interactions followed by inference of gene-regulatory networks. Only one study focused on predicting varying pathway activity using genome-scale metabolic networks. Also, biological network inference studies are only used complementary to functional enrichment techniques (overlap between biological network-based studies and functional enrichment). Among integration-based approaches, most studies fuse single-cell transcriptomes from multiple datasets laterally as compared to vertical fusion of multiple omics data types. Automated gene-prioritization for the identification of AAT targets is the least explored (only 3 studies have attempted prioritization of genes). The bar plot in the bottom left shows comparison of the number of studies which use a particular technique. The bar plots on the top indicate the number of studies that have used a combination of different tools for analysis. The filled dots and lines in the matrix visually represent studies that use different combinations of the tools enlisted in the rows.