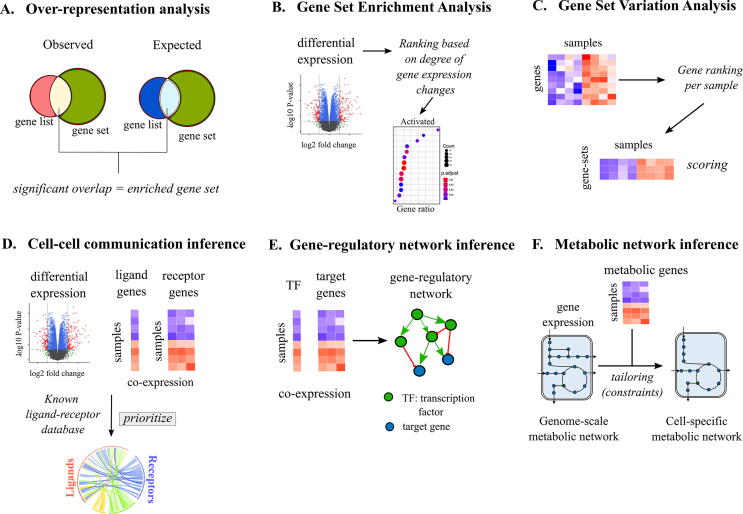
Fig. 2.
Techniques for specialized mechanism discovery. The commonly used tools for mechanism predictions are based either on functional enrichment (A to C) or biological network inference (D to F). (A) Over-representation analysis (ORA): ORA compares the fraction of observed list of genes overlapping with known gene sets (observed) versus the fraction of total list of genes within an organism’s genome that overlaps with known gene sets (expected) to identify enriched gene sets. The overlaps are indicated by Venn diagrams. (B) Gene-set enrichment analysis (GSEA): GSEA ranks genes based on differential expression between control and case samples (indicated by red dots in the Volcano plot) and subsequently, uses the ranks of overlapping genes between the observed and expected cases to score the membership of a gene list to each of the known gene sets (shown as dot plot in the figure). The statistical significance of the enrichment score per gene set is calculated using permutation tests (Box 1). (C) Gene-set variation analysis: GSVA converts the log-normalized gene expression matrix (genes vs samples) into a GSVA score matrix (gene sets vs samples) by ranking genes per sample. (D) Cell-cell communication inference (CCI): CCI methods use the information of differentially expressing (indicated by the red dots in the Volcano plot) or co-expressing ligands and receptors (indicated by heatmap) and compare them with a database of known ligand-receptor interactions to prioritize potential ligand-receptor interactions in a given condition (indicated by a Circos plot connecting ligands to receptors). (E) Gene-regulatory network (GRN) inference: GRN inference methods use the information of transcription factor (TF) expression profile and expression profile of their downstream target genes (indicated by heatmap vectors) to find meaningful co-expressing pairs, which are represented as a network of TF-target interactions. (F) Metabolic network inference: Active, condition-specific metabolic networks are derived by using metabolic gene expression data (heatmap) as biochemical constraints for tailoring a generic genome-scale metabolic network of an organism. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)