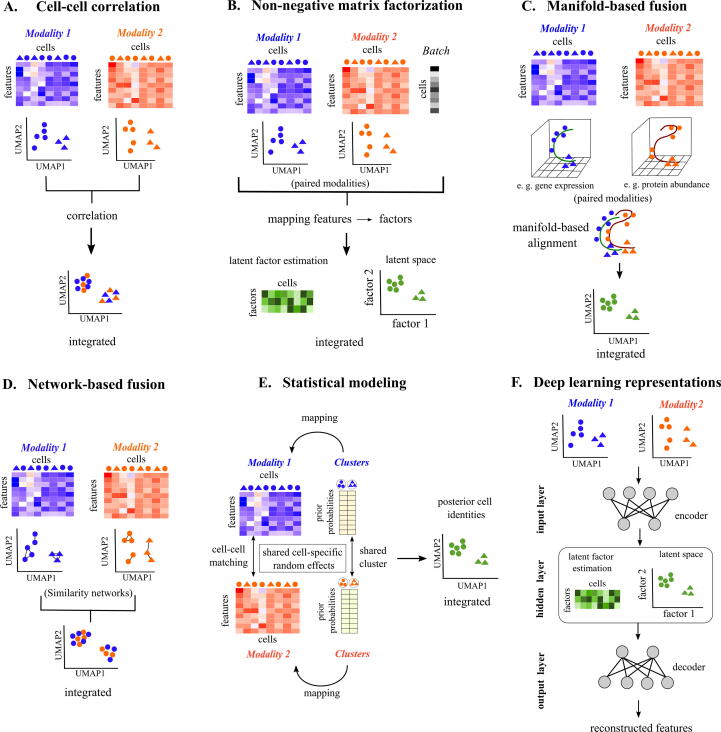
Fig. 3.
Techniques for unsupervised fusion of single-cell multi-omics modalities. In all the figure panels, Modality 1 (red in color), Modality 2 (blue in color) represent two omics modalities. Heatmaps represent variation in feature across cells. Paired modality integrations are illustrated in green color, whereas unpaired modality integration are represented by mixture of blue and orange colors. Colored dots and triangles represent different types of cells. (A) Cell-cell correlation: Cells from modalities 1 and 2 are integrated by measuring correlation between the features from the two omics modalities. (B) Non-negative matrix factorization (NMF): NMF methods map features from two paired modalities and cell-level batch effects to latent factors (Box 1). The number of latent factors being less compared to original number of genes in the figure signifies dimensionality reduction. Cluster identities are assigned to common cells in this latent space (Box 1). (C) Manifold-based fusion: Manifold fusion methods map the input feature dimensions from modalities 1 and 2 to a low-dimensional manifold space (In the figure, 9 row-wise features are mapped to 3 dimensions). The manifolds (Box 1) generated for each paired modality are aligned with each other to identify common cells between modalities. (D) Network-based fusion: Similarity networks are generated for the unpaired modalities 1 and 2. Cells with similar feature profiles are connected to each other within this network. The conserved connections between the two networks are used for integration. (E) Statistical modeling: Statistical modeling methods identify shared clusters and common cells between paired modalities 1 and 2 by generating a probabilistic model (Box 1). As the same prior probability distribution is used for clusters in both modalities to tune the model, shared cell-specific random effects are captured, which are useful for finding posterior cell identities. (F) Deep learning representations: Deep learning for unsupervised omics integration is performed using autoencoders (Box 1), which contains an encoder-decoder scheme. In theory, any of the methods (A to E) can be combined in the hidden layer of the autoencoder scheme to predict cell clusters. Here, the NMF method is shown as an example. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)