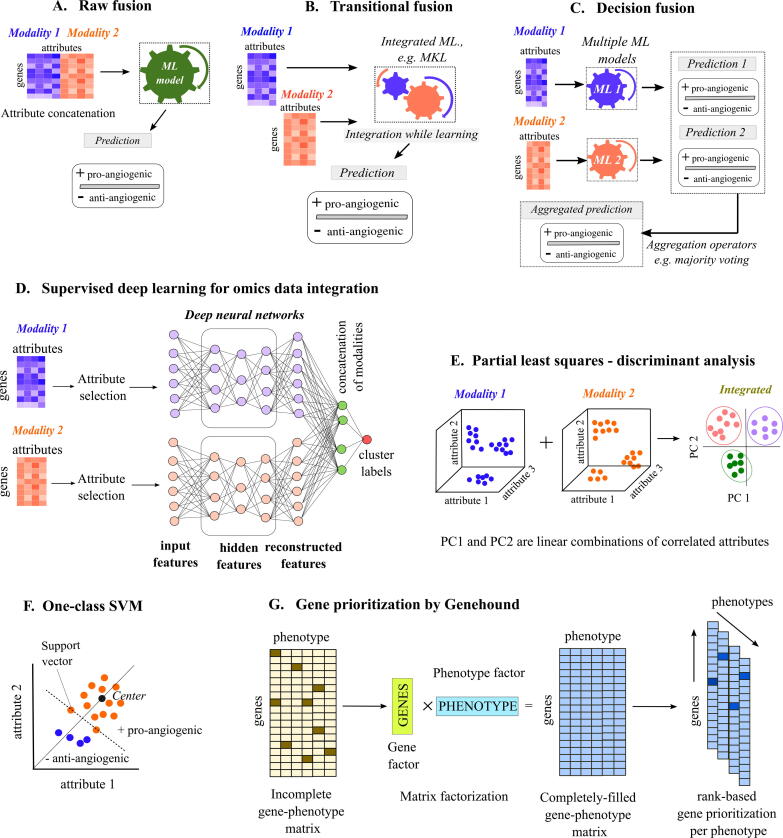
Fig. 4.
Techniques for ML-based supervised fusion of attributes from various data sources. To commonly explain multiple ML techniques, we use a representative example where the aim is to classify genes as pro-angiogenic (+ class) and anti-angiogenic (− class) based on different attributes measured from multiple data sources. (A) Raw fusion: A supervised fusion method that first concatenates attributes from data modalities 1 and 2 (blue and orange colors) and subsequently uses the concatenated dataset for machine learning and classification. (B) Transitional fusion: Here, a structure or pattern is generated for each modalities 1 and 2 separately but they are integrated while learning. The integrated structure is used for classification. (C) Decision fusion: Unlike transitional fusion, the data structures are generated independently for independent learning and only prediction outcomes of + and − class are fused based on majority voting. (D) Supervised deep learning for omics data integration: Deep neural networks (Box 1) are generated for each modality separately. Attributes for each modality are reconstructed an compared with input to evaluate learning performance. The reconstructed features from each omics modality are concatenated finally providing information of cluster labels. (E) Partial least squares-discriminant analysis (PLS-DA): PLS-DA integrates the different attributes from two modalities (blue and orange colors) into PC1 and PC2 and learns the cluster information during integration, and, hence, is an example of intermediate integration. Each PLS-DA component (PC1, PC2) represents a linear combination of correlated attributes from each data source. (F) One-class support vector machine (one-class SVM): Unlike binary SVM (Box 1), in a one-class SVM, different sets of data points are classified into high (large number of points with orange color) or low density regions (low number of points with blue color). The support vectors are then chosen from the high density region depending upon the distance from the center of the high density region to form a hyperplane that is farther from the origin. Based on the labelled information from + pro-angiogenic class, it can predict genes that belong to the - anti-angiogenic class. (G) Gene prioritization by Genehound: Genehound employs a gene prioritization strategy that transforms a gene by phenotype matrix into a completely-filled gene by phenotype matrix using matrix factorization to decompose the gene (green box) and phenotype information (cyan box) as latent factors (Box 1). This completely-filled matrix is used to prioritize genes based on ranking for each phenotype. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)