Figure 1.
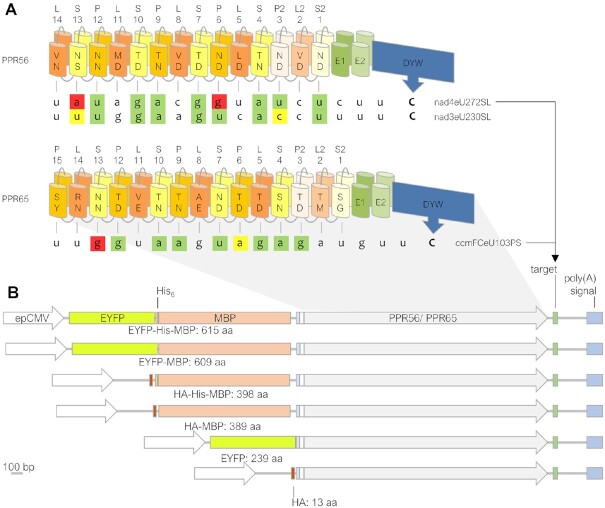
Design of constructs for expressing plant RNA editing factors in human cells. (A) PPR56 and PPR65 are typical pentatricopeptide repeat (PPR) proteins acting as organelle RNA editing factors, featuring a ‘PLS-type’ array of PPRs allowing them to recognize their native targets nad4eU272SL or ccmFCeU103PS, respectively, followed by E1 and E2 extensions and the carboxyterminal DYW cytidine deaminase domain. Positions 5 and L of the P-and S-type PPRs are key positions for RNA binding according to a core PPR-RNA code (24–26) for combinations 5 + L as follows: T/S + N: A, T/S + D: G, N + S: C, N + D: U, N + N: C/U. PPRs are labeled to indicate the respective PPR-type and positions 5 and L with backward numbering starting with S2-1 (here S2-1ND and S2-1SG for PPR56 and PPR65, respectively), as previously suggested (84). Editing sites are labeled with target gene name (nad genes encode for subunits of the NADH ubiquinone oxidoreductase and ccmFC encodes for subunit FC of the cytochrome c maturation machinery) followed by eU, coding sequence position, and resulting amino acid change. Nucleotide shading indicates matches to the corresponding PPR in green, transitions in yellow and mismatches in red. (B) PPR56 and PPR65 were cloned with different combinations of up to three out of four N-terminal tags (EYFP: Enhanced Yellow Fluorescent Protein, His6: 6 x Histidine tag, MBP: Maltose Binding Protein, HA: Hemagglutinin tag). Small grey and white rectangles indicate a TEV recognition site (Tobacco Etch Virus protease) and a short stretch of native editing factor sequence upstream of the first clearly defined PPR, respectively. Protein coding sequences were transcribed from the enhanced Cytomegalovirus promoter (epCMV) together with their respective 46 bp targets cloned downstream followed by the polyadenylation signal.