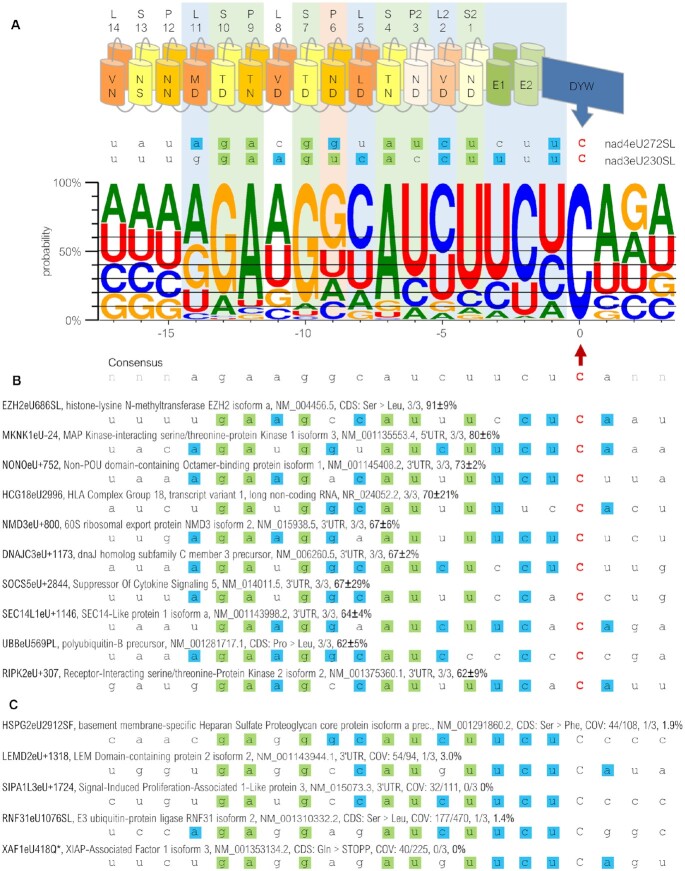
Figure 6.
Off-targets of PPR56 in FACS-sorted IMR-90 cells. (A) Expression of EYFP-PPR56 causes off-target C-to-U RNA editing in the endogenous transcriptome of IMR-90 cells, summarized with a WebLogo (97) created from the sequence environments of 759 edited off-targets (74 putative binding shift candidates were excluded and identical target sequences in different transcript variants of a given gene were only counted once). Targets were juxtaposed with the PPR array of PPR56 aligning the terminal PPR S2-1ND with position −4 upstream of the edited sites (position 0). The horizontal lines indicate arbitrary cut-offs in steps of 10%, starting with 40% for a nucleotide dominating in a given position. Matches according to the established PPR-RNA code (see Figure 1) are shaded green. Evident nucleotide preferences in positions −3, −2 and −1 and in positions −14, −8 and −5 opposite of the L-type PPRs L-11MD, L-5LD and L2-2VD are highlighted by light blue shading. Orange shading of P-6ND highlights its conceptual misfit to a dominating guanidine instead of an expected uridine. (B) A detailed listing of the 10 top-edited off-targets reveals their good matches to the overall consensus profile. Labeling also of the off-target edits uses our proposed nomenclature indicating positions within a coding sequence of the respective gene product and the resulting codon changes with capital letters or with the plus or minus symbol indicating edits in 3’- or 5’-UTRs, respectively (52,98). Description also includes protein name, NCBI accession number, number of replicates with detected editing and average percentage of editing. A complete list of off-targets is available as Supplementary Table S4A. (C) Five exemplary candidate sites are shown, that were not found to be edited or only to very limited degree below our thresholds in single replicates, despite good overall matches to the PPR array of PPR56 (and the consensus off-target profile) in positions −13 to + 1. Coverage of transcripts (COV) in the control sample / in sum of PPR56 replicates is listed in addition. Sites were investigated using IGV Version 2.3.98 (99) for transcriptome analyses.