Figure 7.
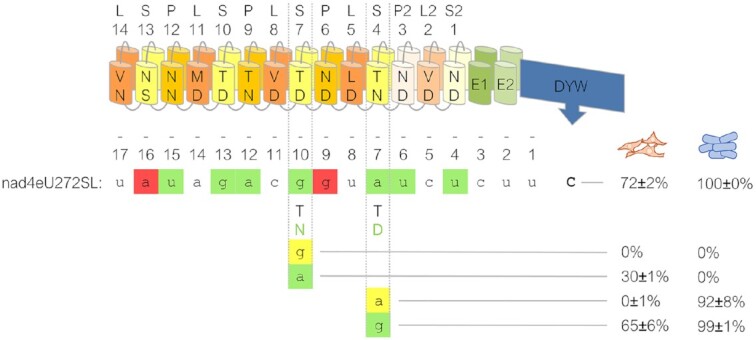
Retargeting of PPR56 by single amino acid exchanges in its PPR array. Single-site mutations were introduced into the ‘Last’ positions of the two PPRs S-7 and S-4 of PPR56 to change their preference for the one vs. the other purine nucleotide: S-7TD > TN and S-4TN > TD changing their (conceptual) preference from G to A and A to G, respectively. A resulting loss of editing at the native nad4 target sequence in the human cells (orange cell icons) in both cases could be compensated by complementing nucleotide changes from G to A in position -10 and from A to G in position -7 upstream of the nad4eU272SL editing site (position 0) resulting in regain of 30% and 65% in the respective transcript populations. The effects of the corresponding mutations observed in the E. coli setup with recombinant His6-MBP-PPR56 are indicated to the right (blue cell icons). Shading of nucleotides is as in Figure 1, scoring of editing efficiencies as in Figure 3.
