Figure 8.
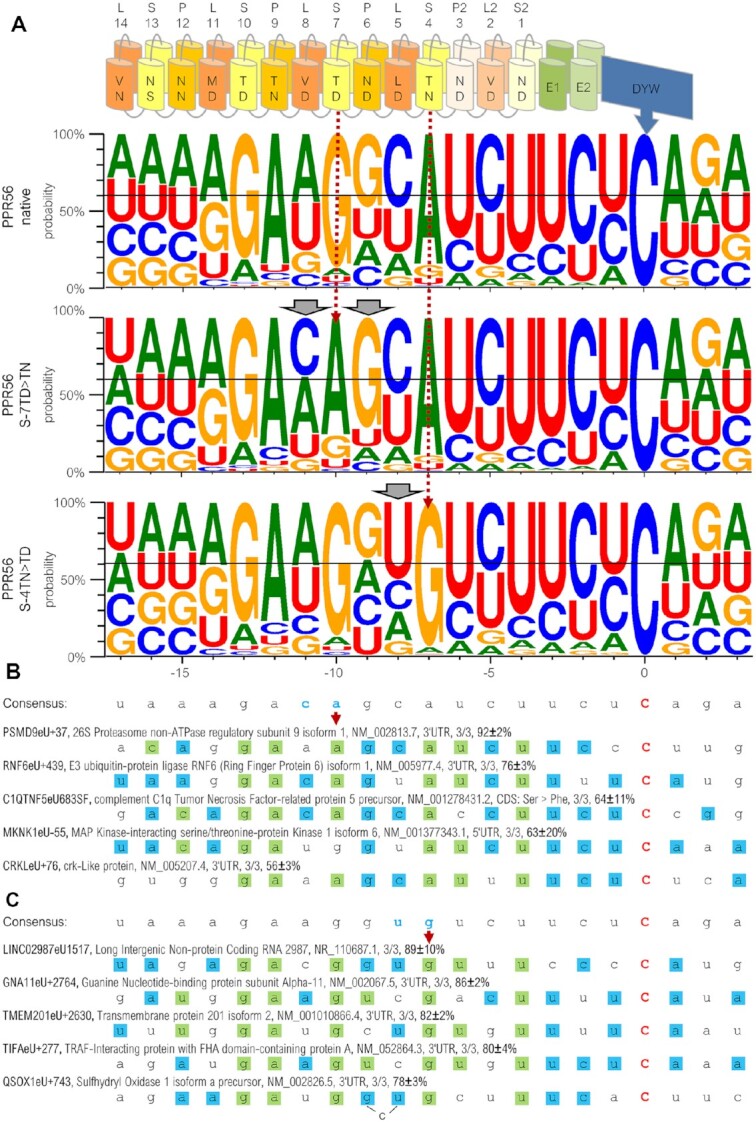
Significant shifts in the off-target sets of two PPR56 mutants. (A) WebLogo profiles (97) are shown for sets of RNA editing off-targets of the native PPR56 (top) and the two PPR mutants S-7TD > TN (middle) and S-4TN > TD (bottom) in IMR-90 cells (for a complete list of off-targets see Supplementary Table S4). Dramatic shifts are identified for the identities of nucleotides in positions −10 and −7 juxtaposed with the mutated PPRs (stippled dark red arrows), as expected. Additional shifts are also seen for neighboring nucleotide identities in positions −11 and −9 for the S-7TN mutant and in position −8 for the S-4TD mutant (grey arrowheads). Five most efficiently edited off-targets each in the analyzed transcriptomes for the S-7TN mutant and the S-4TD mutant are shown in panels B and C, respectively. Labeling includes protein name, NCBI accession number, number of replicates with detected editing and average percentage of editing. A bulged C in position −9 would improve the overall match of the S-4TD mutant to target QSOX1eU + 743.
