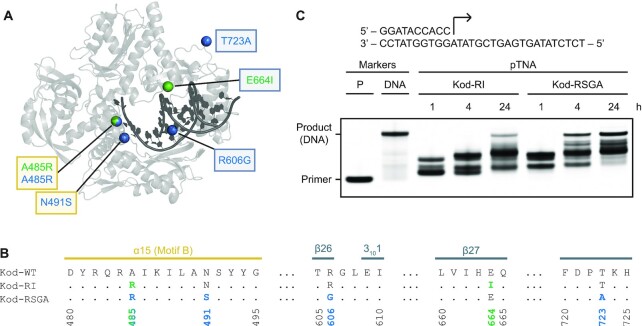
Figure 5.
Engineered mutations in Kod-RSGA. (A) The overall architecture of Kod-RSGA TNA polymerase (gray) bound to a DNA duplex (black) is shown with the positions of the amino acid mutations observed in Kod-RI and Kod-RSGA highlighted as green and blue spheres, respectively. Mutations boxed in yellow and gray occur in the finger and thumb subdomains, respectively. (B) A sequence alignment highlights the secondary structures and positions of the engineered mutations. Both mutations in the finger are part of a conserved sequence known as motif B. (C) A time course demonstrates a faster synthesis rate for the Kod-RSGA. Both enzymes extended a 5′ IR680-labeled DNA primer annealed to a DNA template with ptNDP substrates. A DNA primer (11 nt) and a full-length DNA marker (18 nt) are provided. The reaction contained 0.5 μM IR800-labeled primer–template duplex, 100 μM ptNDPs and 0.5 μM Kod-RI or Kod-RSGA (pre-primed with 1 mM MnCl2), in 1× ThermoPol® buffer for the 15-min intervals at 55°C.