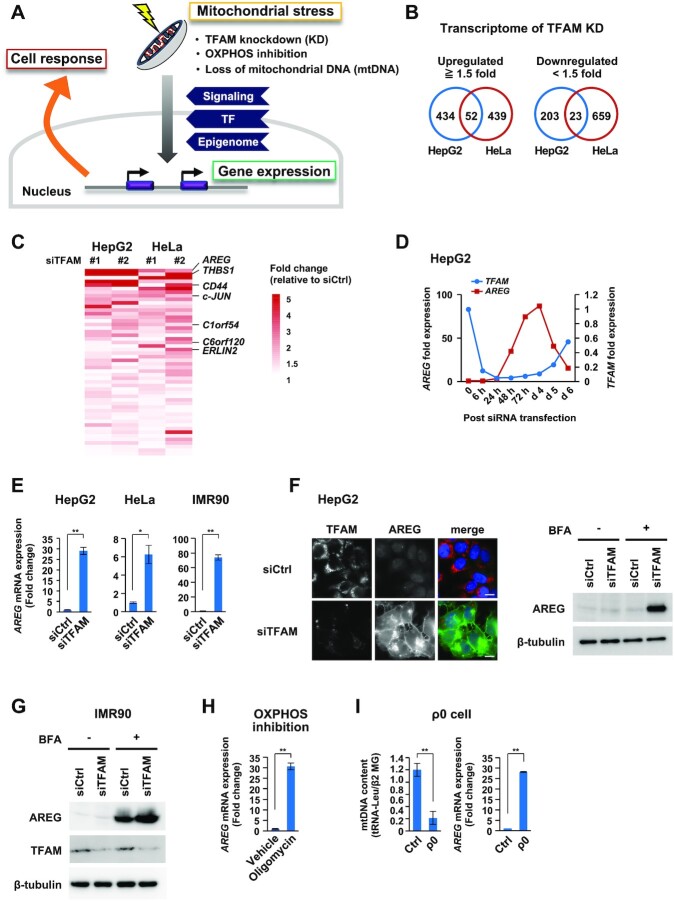
Figure 1.
Transcriptome analyses identify AREG as a mitochondrial stress-induced secretory factor. (A) Study of mitochondrial stress-induced cellular response. Mitochondrial stresses stimulate signaling, transcriptional and epigenomic changes for nuclear gene control. (B) Transcriptome analyses of TFAM knockdown (KD) cells. The Venn diagrams show the number of genes whose expression levels changed by ≥1.5-fold after the distinct TFAM-KD experiments (siTFAM #1 or #2) in HepG2 and HeLa cells. (C) The top-ranked genes commonly upregulated by TFAM-KD include AREG and THBS1 (Supplementary Table S3). Among them, secretory protein genes are shown in the heatmap. (D) Inverse correlation of TFAM and AREG mRNAs after TFAM-KD. RT-qPCR analysis was conducted in the time course. siTFAM #1 was indicated as ‘siTFAM’ in most experiments throughout the study. (E) Induction of AREG mRNA by TFAM-KD. TFAM-KD for 72 h in HepG2, HeLa cells and IMR90 fibroblasts. mRNA levels were normalized by the RPLP0 (ribosomal protein lateral stalk subunit P0) gene and were indicated by mean ± SD (n = 3). **P < 0.01, *P < 0.05. (F) Immunofluorescence and western blot analyses of AREG protein in HepG2 cells. TFAM-KD was carried out for 72 h (left); scale bar: 10 μm. The cells were then treated with 5 μg/ml of brefeldin A (BFA) for 6 h to store intracellular AREG without being secreted (right). (G) Accumulation of AREG in IMR90 cells by TFAM-KD and BFA treatment. (H,I) Induction of AREG mRNA by various mitochondrial stresses. HepG2 treated with 0.5 μg/ml oligomycin for 24 h (H), HepG2-derived ρ0 cells established by ethidium bromide treatment for 10 days (I). mRNA levels were analyzed as described in panel (E).