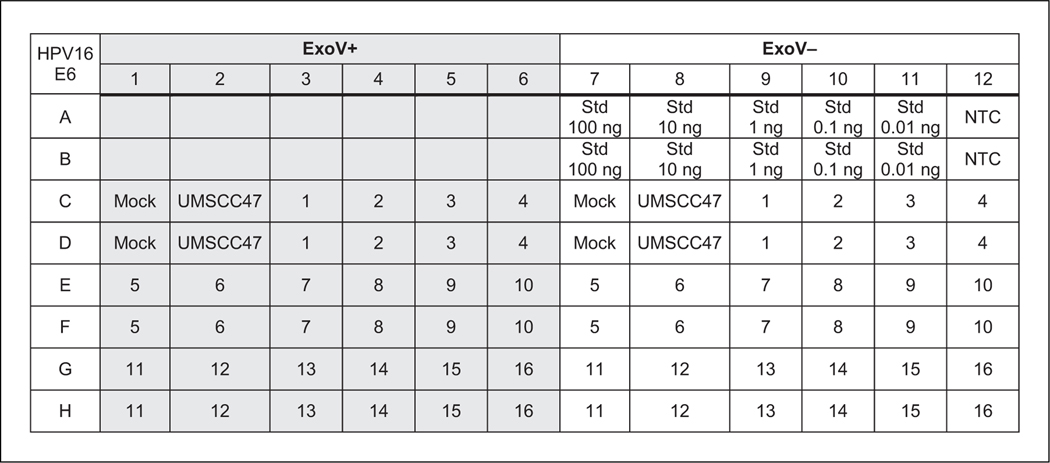
Figure 2.
An example 96-well plate layout using one primer pair in qPCR. This example shows samples and standard curve dilutions in duplicate. ExoV+ labeled wells are samples digested with exonuclease V (ExoV). ExoV– labeled wells are no-enzyme controls to measure the starting input DNA. A total of 16 samples can be analyzed in this setup. Wells labeled Std (standards) represent wells loaded with the UMSCC47 10-fold dilution series. Mock refers to wells with no DNA added to the ExoV+ or Exo– reactions. UMSCC47 DNA monitors ExoV digestion of integrated human papillomavirus (HPV) genomes. NTC represents water as template (a no-template control) for qPCR.