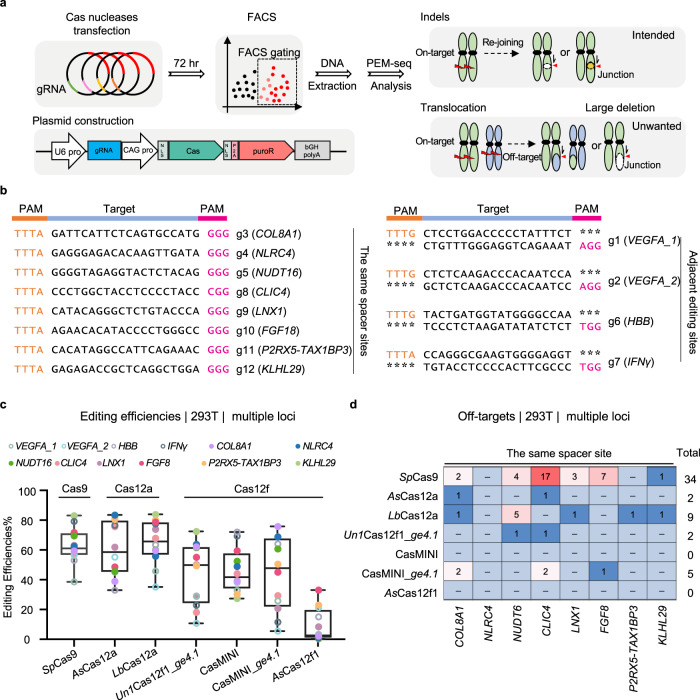
Fig. 2. Evaluation of the global editing outcomes of Cas12f nucleases in parallel with Cas9 and Cas12a as determined by PEM-seq.
a Schematic diagram showing Cas nuclease evaluation by PEM-seq. The plasmids carrying both Cas nuclease-mCherry and single guide (sg) RNA were transfected into HEK293T cells, and the successfully transfected cells were sorted via FACS 72 h post-transfection followed by PEM-seq construction. PEM-seq can simultaneously detect and quantify small indels, large deletions, and chromosomal translocations with off-targets or general double-strand breaks (DSBs). b Detailed guide (g) RNA sequence information of the 12 target sites with PAM sequences for Cas12 and Cas9 in orange or fuchsia, respectively. These target sites were categorized into adjacent editing sites with different gRNA sequences and same-spacer sites with consistent gRNA sequences. c Editing efficiency of the Cas nucleases at the indicated 12 loci as detected by PEM-seq. Editing efficiency indicates the total percentage of insertions, deletions, and translocations. Values from minimum to maximum are shown by the whiskers, and the bounds of the box indicate the first and third quartile (N = 12). The vertical line through the box is the median. Source data are provided as a Source Data file. d Number of off-target editing at the same-spacer sites as detected by PEM-seq methods and identified through the PEM-Q pipeline. “-” means there was no detected off-target site. The indicated locus and Cas nuclease information are marked.