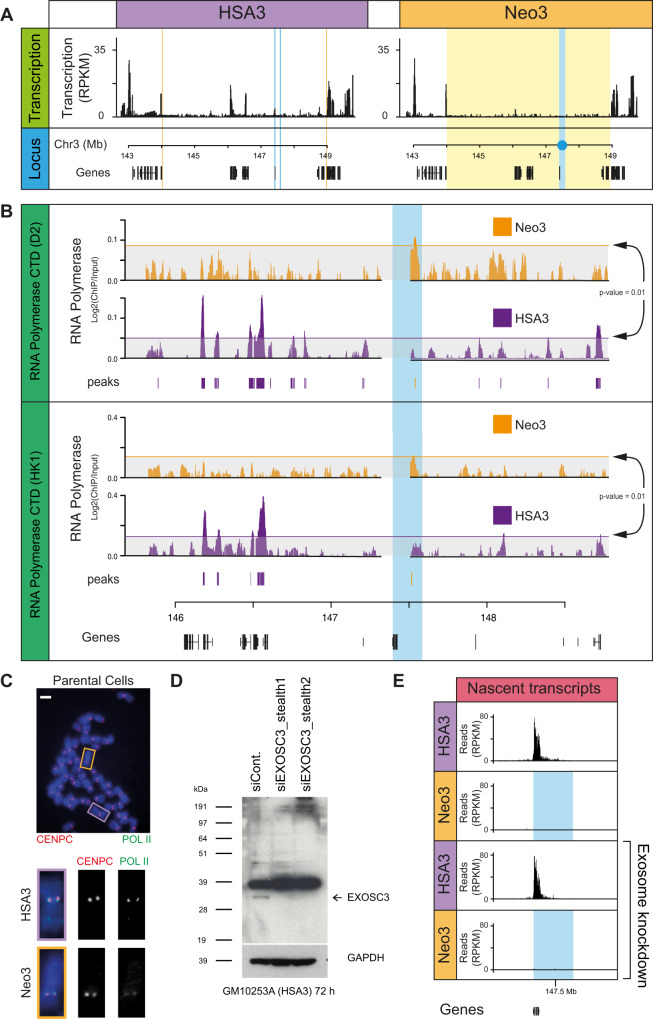
Fig. 2. RNA pol II binding at a functioning human centromere.
A Normalised transcription across HSA3 and Neo3 analysed by RNA-seq. B Distribution of RNA pol II binding at 3q24 for Neo3 (orange) and HSA3 (purple) chromosomes. Top, RNA pol II CTD antibody (D2, Diagenode), bottom, RNA pol II CTD antibody from Hiroshi Kimura (HK1). Horizontal line (arrows) corresponds to a random permutation analysis with p = 0.01. Significant peaks (p < 0.001) called using the Ringo package for Neo3 (orange) and HSA3 (purple) are shown below the tracks. C Representative image of RNA pol II (CTD domain, Diagenode) (green) and CENP-C83 (red) immunofluorescence staining on a metaphase spread from parental cells (HSA3, purple; Neo3, orange) (n = 3 biologically independent experiments). Bar is 5 µm. D Western blot confirming EXOSC3 protein (arrow) knockdown following 72 h RNAi treatment in GM10253A cells (n = 2 biologically independent experiments)). E Distribution of nascent transcripts (normalised RPKM) in 600 kb window around neocentromere (blue), mapped using TT-seq. The top panels correspond to RNAi control, bottom panels are for exosome RNAi knockdown.