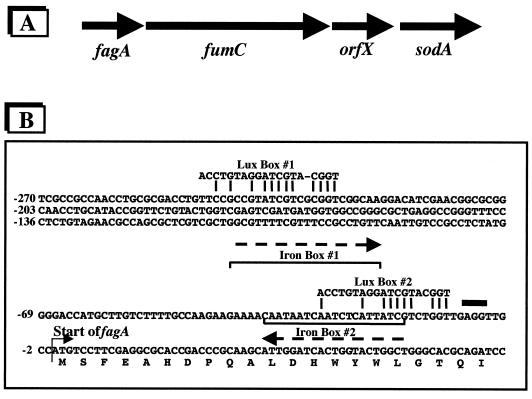
FIG. 6.
A potential mechanism for dual control of sodA by iron-sensitive and QS circuitry. (A) Gene arrangement of the fagA-fumC-orfX-sodA operon. (B) DNA sequence directly upstream of fagA, which is promoter proximal in the depicted operon (24, 25). The Fur regulatory protein (encoded by fur, which is itself regulated by iron availability) utilizes Fe2+ as a corepressor (23). The Fe(II)-Fur complex directly binds to the promoter region of genes that contain a specific regulatory sequence known as the iron box (9). Sequence analysis suggests the presence of iron boxes (orientation of each shown as a dashed line), which is the binding site for Fe(II)-Fur. Note that Mn-SOD production is elevated in fur mutants (25). Iron boxes are located at nucleotide positions -18 to -37 and -21 to -42. Positions of putative Lux boxes, the binding site for the PAI-1–LasR complex, are also shown at -11 to -29 and at -228 to -247. Nucleotide positions that are homologous with the consensus Vibrio Lux box are indicated by the connecting lines. Under high iron conditions, binding of the iron box by the Fe(II)-Fur complex would inhibit the LasR–PAI-1 complex from binding to the Lux box 2 region and would also inhibit transcription possibly originating upstream due to potential activation from binding at Lux box 1. Upon iron starvation, the Fe(II)-Fur complex would not be present, and thus the LasR–PAI-1 complex would be free to activate transcription. The bold thick line indicates a potential ribosome binding site.