Fig. 4. TRIPOD identified trio regulatory relationships in PBMC single-cell multiomic dataset supported by extensive validations.
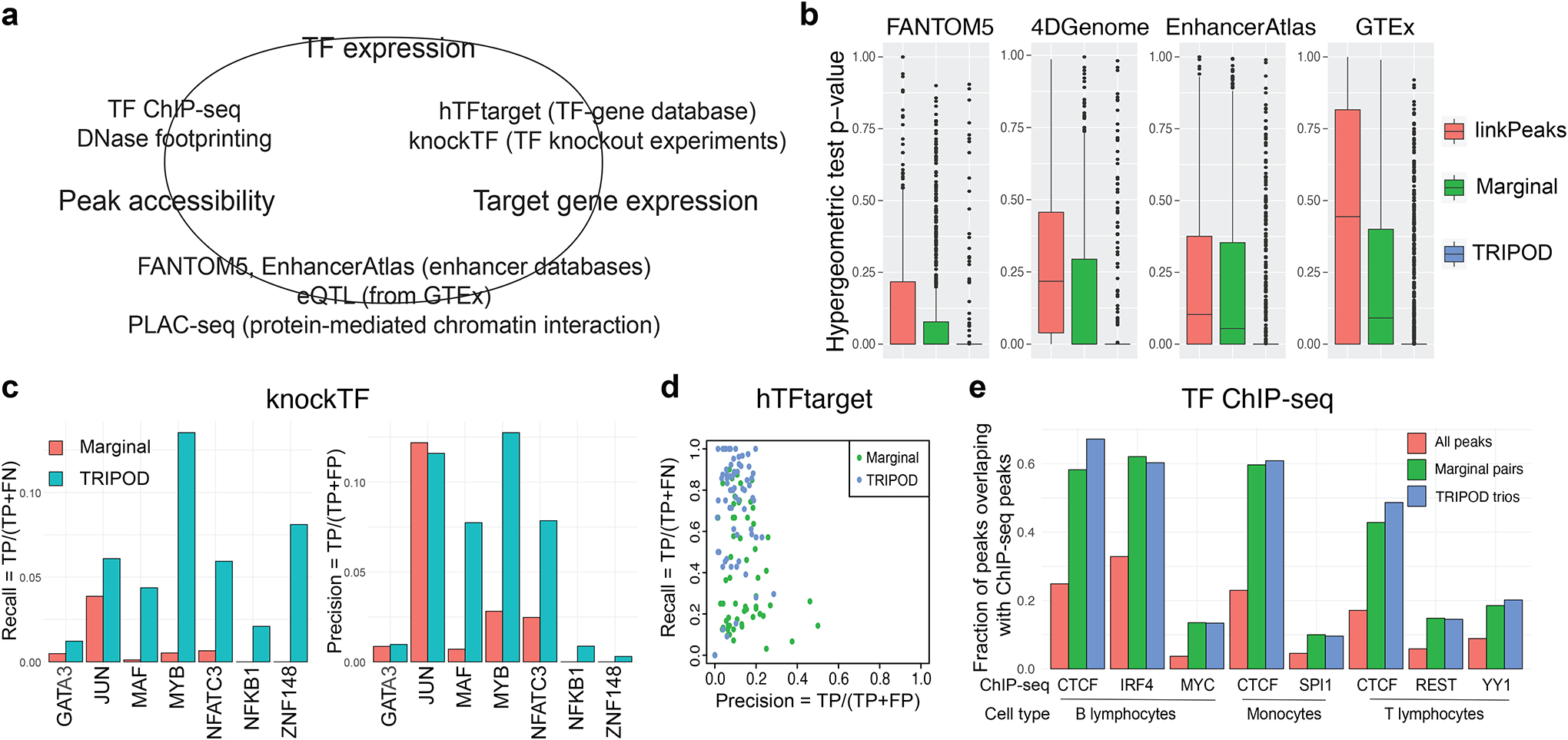
a, A schematic of validation strategies. Shown are external datasets and databases used to validate the links between peak accessibility and target gene expression (peak-gene validation), those between peak accessibility and TF expression (peak-TF validation), and those between TF expression and target gene expression (TF-gene validation). b, Peak-gene validation based on enhancer databases (FANTOM5, 4DGenome, and EnhancerAtlas) and tissue-specific cis-eQTL data from the GTEx Consortium. Box plots show distributions of p-values from gene-specific hypergeometric tests. c, TF-gene validation based on lists of TF-gene pairs from the knockTF database. d, Precision and recall rates for TF-gene pairs using ground truths from the hTFtarget database. e, Peak-TF validation based on eight cell-type-specific TF ChIP-seq datasets (B lymphocytes, monocytes, and T lymphocytes). Fractions of significantly linked peaks and all peaks that overlap with the ChIP-seq peaks are shown.
