Abstract
Medullary thymic epithelial cells (mTECs) clonally delete or divert autoreactive T cells by ectopically expressing a diverse array of peripheral-tissue antigens (PTAs) within the thymus. Although thymic stromal cells with histological features of extra-thymic cell types, like myocytes or neurons, have been observed by light microscopy since the mid-1800s, most modern work on PTA expression has focused on the transcription factor Aire. Here, we highlight recent work that has refocused attention on such “misplaced” thymic cells, referred to collectively as thymic mimetic cells. We review the molecular underpinnings of mimetic cells and their roles in establishing T-cell tolerance, and we propose that mimetic cells play important roles in autoimmunity. Finally, we suggest future directions for this emerging area.
Peripheral-tissue antigen expression in thymic epithelial cells
All stages of pre-immune T-cell maturation in vertebrates, including T-cell lineage commitment, T-cell receptor (TCR) formation, CD4+ vs CD8+ lineage choice, positive selection, negative selection, and agonist selection (see Glossary), occur within the thymus, with thymic epithelial cells (TECs) playing key instructive roles throughout (reviewed in [1]). In the cortex, cortical TECs (cTECs) provide signals for lineage choice and major histocompatibility complex (MHC) restriction, while in the medulla, medullary TECs (mTECs) express peripheral-tissue antigens (PTAs) to preview the peripheral self to maturing thymocytes. The latter process allows for identification of T cells bearing autoreactive TCRs and their clonal deletion or diversion into the regulatory T cell (Treg) lineage, thereby enforcing central tolerance.
Since the original observation of PTA expression by mTECs [2-4], the molecular basis of this unusual phenomenon has received considerable attention. Some early models postulated a relationship to histologically distinct epithelial cells that had been observed in the medulla since the mid-1800s [5], long before the role of the thymus or even basic principles of adaptive immunity were understood. These “misplaced” cells included skin-like Hassall’s corpuscles, lung-like ciliated cysts, and muscle-like myoid cells, to name a few (see Box 1) [6-8]. However, the transcription factor (TF) Aire was soon implicated as a major regulator of PTA expression, with population-level transcriptomic analyses of mTECs from Aire−/− mice showing decreased expression of most PTAs [9]. Consistent with a major role for Aire in central tolerance, mice and humans with mutations in the Aire/AIRE gene developed autoantibodies against Aire-dependent antigens and multiorgan lymphocytic infiltration [3,9-13]. These results shifted the focus of studies on PTA expression to Aire-dependent mechanisms, and interest in the early histologic observations waned.
BOX 1: Mimetic cell types.
Basal (skin/lung) mTEC:
mimetic cells (MC) resembling basal cells of the skin and lung that putatively give rise, as in the periphery, to more terminally differentiated MC, such as keratinocyte mTECs and secretory/ionocyte mTECs [38].
Enterocyte/hepatocyte mTEC:
Hnf4a+ Hnf4g+ MC expressing transcripts associated with gut enterocytes and liver hepatocytes, such as Vil1, Aldob and Apoa4 [38]. These cells putatively give rise to microfold mTECs [38].
Ciliated mTEC:
Foxj1+ Rfx2+ Trp73+ MC exhibiting polarized cilia, line respiratory cysts in the thymus, and expressing transcripts associated with ciliated cells, such as Dynlrb2, Pifo and Tubb4b [7,28,34,38]. Expression of a model antigen in ciliated mTECs is sufficient to induce cognate T-cell tolerance [38].
Ionocyte mTEC:
Foxi1+ Foxi2+ MC expressing transcripts (Cftr, Slc12a2, Atp6v1b1) associated with ionocytes, and ion-channel-rich cells found in the kidney and lung epithelium [37,38].
Keratinocyte mTEC:
Grhl1+ MC morphologically resembling skin keratinocytes, producing microscopically detectable Krt10+ cornified bodies that in humans have long been known as Hassall’s corpuscles [6,33,38].
Microfold mTEC:
Spib+ Sox8+ MC morphologically resembling Peyer’s patch microfold cells, with dendritic processes and an associated “lymphocyte pocket.” They express transcripts associated with microfold cells, such as Ccl6, Tnfrsf11b, and Gp2 [38]. Microfold mTECs are lost in Spib−/− and Sox8−/− mice [38], and the differentiation of thymic B cells is perturbed [38,39].
Muscle mTEC:
Myog+ MC morphologically resembling and expressing transcripts (Ckm, Des, Myl1) associated with skeletal muscle [8,36,38]. Expression of a model antigen in muscle mTECs is sufficient to induce cognate T-cell tolerance [38].
Neuroendocrine mTEC:
Foxa2+ Foxa3+ Insm1+ Ascl1+ MC expressing transcripts encoding neuroendocrine markers (Scg5, Snap25, Chga, Stxbp5l) and possessing abundant secretory granules [35,38,39]. Under the umbrella of neuroendocrine mTECs, further heterogeneity exists including Ptf1a+ pancreatic-like, Cdx2+ enteroendocrine-like, Pax6+, and Sox11+ subsets ([38] and our unpublished analysis of data from [38]). Neuroendocrine mTECs are lost in Foxn1cre Insm1flox and Foxn1cre Ascl1flox mice, and Foxn1cre Insm1flox develop autoantibodies against the thyroid and enteroendocrine-cell-rich gastric fundus [39].
Parathyroid mTEC:
Gcm2+ MC expressing microscopically detectable parathyroid hormone (PTH) [29,58]. Whether these cells are bona fide thymic epithelium or ectopic developmental remnants is controversial and has been debated elsewhere [29,58,59].
Secretory mTEC:
Foxa1+Spdef+ MC expressing transcripts (Gabrp, Aqp4, Scgb3a2, Sftpd, Muc5ac, Muc5b) associated with mixed secretory cell types, including goblet cells, club cells, and alveolar epithelial cells [38].
Thyroid mTEC:
Poorly characterized MC identified as thyroglobulin- and calcitonin-expressing cells by immunofluorescent microscopy [29] More work is needed to determine whether these cells are regular features of the MC compartment.
Tuft mTEC:
Abundant Pou2f3+ MC expressing markers of tuft cells (IL-25, ChAT, Dclk1), mediate tolerance to tuft-cell-restricted antigens and control the accumulation of type 2 ILCs and NKT cells in the thymus [32,33,60]. Tuft mTECs are lost in Pou2f3−/− and Trpm5−/− mice [32,33].
Here, we briefly review early studies on PTA expression and Aire before highlighting recent work that has refocused attention on “misplaced” thymic stromal cells, collectively termed mimetic cells. We discuss molecular, cellular, and immunological aspects of mimetic cells, and argue that mimetic cells likely play important roles in the pathogenesis of autoimmunity. Finally, we suggest future directions for this emerging area.
The Aire model of PTA induction
While compelling in many respects, the Aire model of PTA induction left several questions unanswered. First, Aire has little-to-no sequence-specific binding activity [14]; how, then, could it select genes to induce, especially given the diverse and disparate nature of its targeted genes? Second, the expression of many PTAs is diminished, but not extinguished, in Aire−/− mTECs [15,16]; might Aire act as an amplifier of PTA expression, rather than its primary inducer? Finally, individual PTAs, far from being expressed in all mTECs, are instead expressed in a variegated fashion, with any given PTA typically expressed in only 1-5% of mTECs [4]; why would Aire, which is uniformly expressed in a major fraction of mTECs, induce PTAs so heterogeneously?
Especially given this last point, single-cell transcriptomics were intuitively appealing to study the landscape and mechanism of PTA expression. Early work in this area used single-cell reverse transcription and polymerase chain reaction (RT-PCR) and plate-based single-cell RNA sequencing (scRNA-seq) to examine PTA coexpression patterns in murine and human mTECs [16-20]. In general, though, these studies were limited by throughput constraints and, although some non-randomness in PTA expression was observed, few coherent patterns emerged, leading to the consensus that PTA expression in mTECs was “quasi-random” and proceeded with “ordered stochasticity.” Contemporaneously, several molecular studies characterized Aire’s influence on gene regulatory mechanisms, such as RNA polymerase II pausing, epigenetic modifications, RNA splicing, super-enhancer activity, and chromatin looping [16,21-26]. From this work, the idea emerged that Aire could repurpose general transcriptional mechanisms to induce tissue-specific genes.
Defining mimetic cells
In parallel with these Aire studies, a few investigators continued to study the “misplaced” cells of the thymic medulla. Several microscopic and RT-PCR-based studies showed that the murine thymus contains clusters of cells resembling lung, thyroid, and parathyroid epithelia [27-30], raising the possibility that mTECs might appropriate the developmental programs of peripheral tissues for tolerance. However, no evidence of a functional role for these cells in tolerance was provided, and a subsequent study found that TEC-specific conditional deletion (Foxn1cre) of Pdx1, a pancreatic lineage-defining TF, did not affect mTEC expression of pancreatic genes [31]. Meanwhile, contemporaneous work on Aire continued to confirm its importance in PTA induction, eventually leading to a widespread consensus that Aire, rather than appropriated developmental mechanisms, was the major driver of PTA induction.
The tide shifted with the advent of higher-throughput scRNA-seq, which uncovered rare transcriptomic equivalents of histologically defined mTECs, including ciliated, myoid, and tuft mTECs [32-37]. These findings once again foregrounded the possibility of compartmentalized, non-random PTA expression. Single-cell characterizations of tuft mTECs were especially prescient, as the combined observations of two studies went beyond phenomenology to show that tuft mTECs relied on a tuft-cell lineage-defining TF, Pou2f3, for their accumulation, could mediate tolerance to a tuft-cell-restricted antigen, and had influences on thymic cell subsets beyond conventional αβ T cells [32,33].
Recently, based on the single-cell assay for transposase-accessible chromatin and sequencing (scATAC-seq), our own study of chromatin accessibility in individual mTECs systematized these individual cell-type observations to describe a whole constellation of peripheral-cell-mimicking mTECs with principled differentiation and PTA expression [38]. We found that the accessible chromatin of mTEC subsets was enriched for the signatures of lineage-defining TFs from peripheral cell types, and we hypothesized that these TFs might be co-opted by mTECs to drive expression of lineage-specific gene programs within the thymus. Indeed, targeted scRNA-seq of mTECs found numerous mTEC subtypes expressing paired lineage-defining TFs and lineage-specific programs, such as mTECs resembling Grhl+ keratinocytes, Foxa+ neuroendocrine cells, Foxj1+ ciliated cells, Hnf4+ enterocytes/hepatocytes, Spib+ Sox8+ microfold (M) cells, Foxi+ ionocytes, Myog+ skeletal muscle, Spdef+ goblet cells, and Ptf1a+ pancreatic secretory cells[38]. Many subtypes corresponded to mTECs that had been phenomenologically described by histology or scRNA-seq [6-8,34-37], and, as we sequenced more cells, more subtypes continued to emerge. Of note, another preliminary study, recently reported in preprint, found a similar diversity of mTECs using paired scATAC-seq and scRNA-seq [39]. Collectively, we use the term “mimetic cells” to refer to these mTECs with the lineage-defining TFs, chromatin landscapes, and gene expression programs of peripheral cell types (Box 1).
Principles of mimetic cells
Synthesis of our study and others has revealed some general principles of mimetic cell biology. Most fundamentally, mimetic cells have biologically logical PTA expression patterns controlled by lineage-defining TFs. Lineage-defining TFs correlate with ectopic expression of lineage-restricted programs within mimetic cells [38,39]; lineage-defining TFs bind specifically to the accessible chromatin of their corresponding mimetic cells [38]; and TEC-intrinsic loss of lineage-defining TFs results in loss of the corresponding mimetic cells, as shown by thymic graft experiments with Spib−/− or Sox8−/− murine thymi lacking microfold TFs [38] and by TEC-specific conditional deletion (Foxn1cre) of the neuroendocrine TFs Insm1 or Ascl1 [39].
As concerns the provenance of mimetic cells, lineage tracing with AireCreERT2 and CsnbCre mice has shown that most mimetic cells (excepting muscle mTECs) differentiate from Aire-expressing progenitors [33,38-40]. Importantly, however, differentiation of mimetic cells downstream of Aire does not imply that they require Aire for their differentiation. On the contrary, many mimetic cell types are numerically reduced, but not absent, in Aire−/− mice, giving a satisfying explanation for why, in population-level transcriptomic studies of mTECs, loss of Aire diminishes, but does not extinguish, the expression of many PTAs [15,33,38,39]. Aire does not appear to directly transactivate mimetic-cell PTAs, as gene expression within mimetic cells is largely unperturbed in Aire−/− mice [38,39], and Aire is not strictly required for binding of lineage-defining TFs to mimetic-cell chromatin, though it does enhance the binding of some TFs, such as Grhl1 and Pou2f3 [38].
To what extent do mimetic cells transdifferentiate into their peripheral counterparts? Many mimetic cells take on morphological features of their counterparts, such as polarized cilia in ciliated mTECs, lymphocyte pockets in microfold mTECs, and apical tufting in tuft mTECs [27,32,33,38]. Nonetheless, RNA-seq of mimetic cells and their peripheral counterparts show that mimetic cells retain the mTEC gene program as their core identity, with their cell-type-specific programs layered on as a minor (albeit substantial) fraction of their transcriptomes [33,38]. Spatially, immunofluorescent microscopy of murine and human thymi has shown that some mimetic cells (e.g., microfold and muscle mTECs) are scattered evenly through the thymic medulla, whereas others (e.g., ciliated, keratinocyte, and tuft mTECs) form coordinated microstructures such as respiratory cysts and Hassall’s corpuscles [33,34,38]. A random distribution of mimetic cells would seem to be most efficient for negative selection by maximizing the likelihood of a thymocyte encountering its cognate antigen, but mimetic-cell microstructures may also serve as unique niches for specific thymic processes: for example, histologic analysis and in vitro co-culture experiments have suggested that human Hassall’s corpuscles facilitate Treg induction by dendritic cells (DCs) [41].
Finally, mimetic cells have been shown in specific instances to be necessary and sufficient for antigen-specific T-cell tolerance. Transplantation of Pou2f3−/− thymi into nude mice led to the development of autoantibodies against the tuft-specific antigen, IL-25, and conditional deletion of Insm1 and Ascl1 in mTECs led to the development of autoantibodies against the thyroid and the enteroendocrine-rich gastric fundus [39], demonstrating the necessity of mimetic cells for tolerance [33]. Conversely, induced expression of a model self-antigen, yellow fluorescent protein (YFP), in ciliated and muscle mTECs using lineage-specific Cre drivers (Foxj1Cre, Ckmcre) diminished the number of YFP-reactive T cells in the periphery, indicating that self-antigen expression in mimetic cells suffices for tolerance [38]. Extrapolating from these data, we argue that mimetic cells are likely to be the primary drivers of thymic tolerance for PTAs whose expression is restricted to mimetic cells.
An integrated model of PTA expression
The newly recognized role of mimetic cells should not be taken to negate the well-established importance of Aire in central tolerance. Rather, the two mechanisms require integration to provide a more comprehensive view of thymic selection. Several key insights into Aire function, gleaned from our recent study and elsewhere, inform on its role in our integrated model. First, not surprisingly, Aire exerts its direct effects where it is expressed, within Aire-stage mTECs, which have a chromatin and transcriptional state distinct from that of mimetic cells [38]. At the chromatin level, Aire enhances accessibility at Aire-binding sites and Aire-induced genes, without altering accessibility at Aire-neutral genes or at active or repressive histone marks more broadly [25,38]. Notably, these results are not in agreement with the recently proposed notion, based on bulk ATAC-seq data, that Aire acts primarily as a repressor, an interpretation that we suggest was confounded by bulk amalgamation of primary and secondary effects of Aire on mTEC chromatin [42]. At a transcriptional level, Aire induces diverse PTAs, unique to Aire-stage mTECs, and with a predilection for genes encoding inflammatory/antimicrobial peptides such as S100a8, S100a9, and Defb19, as well as neuropeptides such as Ins2, Gip, and Ppy [38,39].
Altogether, we envision a model (Figure 1, Key Figure) in which transit-amplifying TECs differentiate into Aire-stage mTECs, which express high amounts of MHC class II molecules and a select set of PTAs that are directly induced by Aire through repurposing of general transcriptional mechanisms and cooperation with pre-expressed mTEC TFs. At some subsequent point, diverse lineage-defining TFs are induced by Aire, spatial cues, and/or other signals. Once expressed, lineage-defining TFs drive the differentiation of diverse mimetic cell types, producing and maintaining their chromatin states and transcriptional programs, including mimetic-cell-specific PTAs. Maturing autoreactive thymocytes can undergo negative or agonist selection against the full range of PTAs expressed by Aire-stage mTECs and mimetic cells, which collectively encompass what were previously thought of as “Aire-induced” PTAs (Figure 2). Finally, defects in the thymic action of Aire or specific TFs lead to specific PTA deficiencies, giving rise to specific syndromic manifestations of autoimmunity.
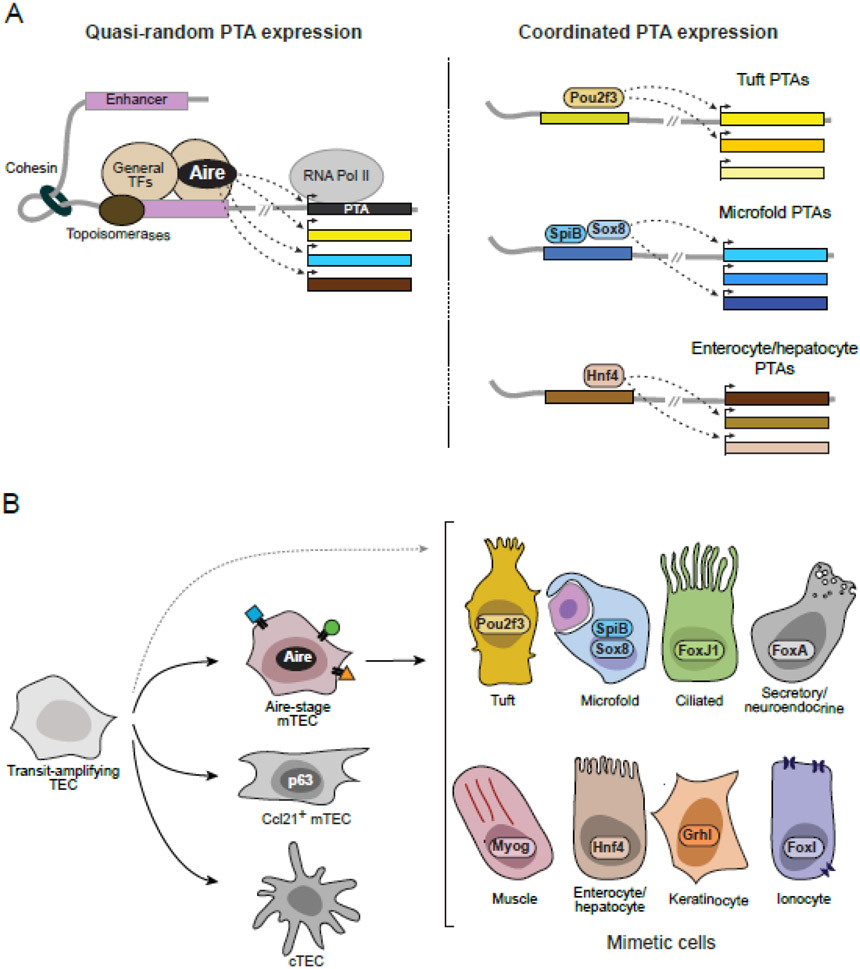
Figure 1 (Key Figure): Model of thymic PTA expression.
(A) Molecular model of PTA induction by Aire (left) and lineage-defining TFs (right). Aire binds promiscuously to mTEC enhancers and promoters and interacts with general transcriptional mechanisms to quasi-randomly activate expression of diverse PTAs [25]. In contrast, lineage-defining TFs bind specifically to cell-type-specific enhancers and drive coordinated expression of biologically coherent sets of PTAs [38]. (B) Cellular model of PTA expression by mTECs. Transit-amplifying TECs give rise to multiple TEC lineages, including cTECs, Ccl21+ mTECs, and Aire-stage mTECs [40]. Aire-stage mTECs further differentiate into diverse mimetic cell types characterized by the lineage-defining TFs, chromatin landscapes, and gene expression programs of peripheral cell types [38,39]. Some mimetic cells (i.e., muscle, tuft mTECs) may not necessarily go through an Aire-positive mTEC stage, as indicated by the dashed line [33,38,39]. Note that Ccl21+ mTECs have been called “immature” mTECs, but likely represent a separate mTEC lineage as opposed to an “immature” progenitor [40].
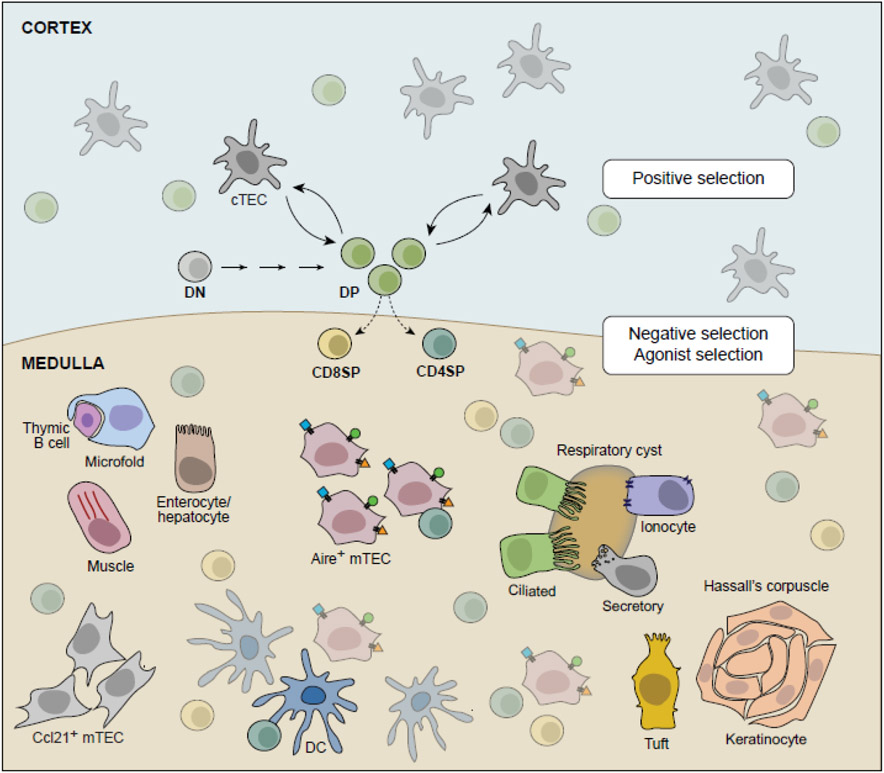
Figure 2: Model of αβ T-cell selection.
Simplified model of αβ T-cell selection. Most thymocytes mature from CD4−CD8− double-negative (DN) precursors into CD4+CD8+ double-positive (DP) thymocytes expressing rearranged αβ TCRs. DP thymocytes undergo positive selection for self-MHC molecule restriction through interactions with cTECs; surviving thymocytes become CD4+ and CD8+ single-positive (SP) thymocytes and undergo negative selection or agonist selection through interactions with thymic APCs in the cortex (ubiquitous antigens) and medulla (PTAs). mTECs, including diverse mimetic cells, provide PTAs for negative and agonist selection, either directly through presentation on MHC molecules or indirectly through antigen transfer to other thymic APCs, including dendritic cells (DCs).
Future perspectives
Many new questions have arisen (see Outstanding Questions). Some key areas of future work include comprehensively cataloging and characterizing mimetic cells, unraveling the molecular relationship between lineage-defining TFs and Aire, elucidating the impacts of mimetic cells on other cell types, and testing the role of mimetic cells in mouse and human autoimmunity.
OUTSTANDING QUESTIONS BOX.
Precisely which TFs control which mimetic cells?
How do Aire and lineage-defining TFs integrate, or not, to induce PTAs and mimetic cells?
Can TF networks be dynamically controlled to output different mimetic cells?
What makes mTECs permissive to the ectopic action of lineage-defining TFs?
Are there unique advantages to expressing PTAs in the context of mimetic cells?
What impact do mimetic cells have beyond selecting the αβ T-cell repertoire?
What are the immunological sequelae of defects in the various mimetic cell subtypes?
How does variation in human mimetic cells contribute to autoimmune disease risk?
Comprehensively characterizing mimetic cells
Most fundamentally, we need a comprehensive understanding of every mimetic cell type: their progenitors, relative abundances, associated PTAs, driving TFs, proliferative potential, capacity to transdifferentiate into other mimetic cell types, effects on T cells and other cells, and immunological consequences when deleted or defective. This last point is of particular interest as the consequences of mimetic cell dysfunction may inform a new understanding of autoimmune disease. For many of these experiments, new surface markers and genetic tools will need to be developed to permit facile analysis of the diverse mimetic cell types.
Molecular regulation of PTA expression
From a molecular perspective, we await a fuller picture of how Aire and lineage-defining TFs collaborate, or not, on mTEC chromatin to present a full diversity of PTAs to maturing thymocytes. Biochemically, Aire has chromatin-binding, rather than DNA-binding properties, suggesting that it may function more as a coactivator than a traditional TF [14,43-45]. Future experiments should explore whether Aire truly “chooses” its genes, or whether it is steered in all cases by sequence-specific TFs and/or site-specific signals. The chromatin of mTECs also seems uniquely permissive to the action of Aire and lineage-defining TFs, a principle established by early experiments showing stronger effects of Aire on gene expression in mTECs compared to other cell types and now underscored by observations of mTEC transdifferentiation into mimetic cells [38,46]. Comparative studies of mTECs, stem cells, and peripheral epithelia may reveal cis- and trans-regulatory features that permit the induction of a diverse repertoire of PTAs in mTECs while restraining the wholesale conversion of mTECs into bona fide tissues or teratomas.
Cellular functions of mimetic cells
At the cellular level, Aire-stage mTECs and mimetic cells represent clearly distinct cell states, raising the question of what the advantage of two mechanisms of PTA expression might be. One idea is that Aire-stage mTECs express a broad swath of PTAs but may not efficiently express inflammatory or other environmentally induced pathways for tolerization, whereas mimetic cells may coordinately induce pathways associated with their respective cell types, such as type 2 alarmins made by tuft cells, and Peyer’s patch chemokines made by microfold cells [38]. Additionally, and not mutually exclusively, antigens derived from Aire-stage mTECs and mimetic cells may differ in their modes of presentation, thereby diversifying the peptide pool available for tolerance (i.e, differential peptide processing, differential use of MHC molecules, direct vs indirect presentation). Such a mechanism might be akin to the function of the thymoproteasome in cTECs [47]. These hypothetical possibilities should be assessed experimentally for their importance in central tolerance induction.
Beyond providing PTAs to conventional αβ T cells, mimetic cells may also play other roles in thymus biology. Hints of such roles have already emerged: Pou2f3−/− thymi lacking tuft mTECs show increased numbers of innate lymphoid group cells (ILC) but decreased NKT cells [32,33], and Spib−/− and Sox8−/− thymi lacking microfold mTECs show increased numbers of thymic B cells, but impaired generation of thymic IgA+ plasma cells [38,39]. Many mimetic cell types express transcripts encoding molecules that may influence their surrounding milieu, such as Il10 and Il25 by tuft mTECs and Ccl6, Ccl20, and Tnfrsf11b by microfold mTECs [32,33,38,39]. Alongside characterizations of conventional αβ T-cell repertoires, then, mice with defects in mimetic cells should also be evaluated for any impacts on unconventional T cells and other thymic cell subsets.
Mimetic cells and autoimmunity
While nearly all work to date on mimetic cells has been in mice, the coordinated provision of PTAs by mimetic cells has important implications for our understanding of autoimmune syndromes in humans. Much basic science work has shown that defects in central tolerance can causally cascade into autoimmunity, such as classic studies showing that variation in the number of tandem repeats at the insulin promoter can diminish thymic insulin expression and central tolerance to insulin, resulting in anti-insulin T cell and autoantibody responses and ultimately type 1 diabetes [48-50]. Clinically, however, most polygenic autoimmune diseases are still treated as phenomenological syndromes rather than as rooted in causal molecular defects, at least in part because few theoretical frameworks to understand autoimmune risk have emerged beyond polymorphism at human leukocyte antigen (HLA) loci.
We propose that mimetic cells may provide exactly such a framework: defects in the thymic action of lineage-defining TFs would lead to loss of tolerance to biologically coherent sets of antigens, resulting in biologically coherent patterns of autoimmunity. Indeed, lineage-defining TFs from mimetic cells have already been identified as risk loci in many human autoimmune diseases, including HNF4A in inflammatory bowel disease, SPIB in primary biliary cirrhosis, and SOX8 in multiple sclerosis [51-53]. Moreover, mounting evidence suggests that HLA-linked autoimmune risk operates in large part within the thymus, during the establishment of central tolerance, providing support in favor of a “central hypothesis” for autoimmunity [54-57]. Human mimetic cells, some subsets of which have already been observed [36-38], should be evaluated for their adherence to the principles established in mouse and to test the hypothesis that mimetic cells are compromised in certain autoimmune syndromes.
Concluding remarks
The discovery of a constellation of mimetic cells in the thymus has substantially widened our view of mTEC biology and T-cell tolerance; nonetheless, we still have much to learn about these “tolerogenic masqueraders” (see Outstanding Questions). We now understand that Aire and lineage-defining TFs work together to induce PTAs in a principled and biologically logical fashion, as opposed to quasi-random induction by Aire alone. Moreover, proof-of-principle experiments have demonstrated that mimetic cells are important for self-tolerization of maturing thymocytes. Comprehensive characterizations of mimetic cells and their relationships to other thymic cell types can help reveal the influence of mimetic cells on the immune system and their importance in controlling autoimmunity. Studies of this remarkable phenomenon may also yield new insights into transcriptional regulation and developmental biology more generally.
SIGNIFICANCE.
Centuries-old observations of “misplaced” cells in the thymic medulla have recently been unified in the description of mimetic cells, specialized thymic epithelial cells that appropriate the lineage-defining transcription factors of diverse cell types to express peripheral-tissue antigens within the thymus and tolerize maturing T cells. Defects in mimetic cells may play major roles in the pathogenesis of autoimmunity.
HIGHLIGHTS.
Mimetic cells are medullary thymic epithelial cells that mimic diverse extra-thymic cell types
Lineage-defining transcription factors drive peripheral mimicry in mimetic cells
Mimetic cells can be necessary and sufficient for antigen-specific T-cell tolerance
Compartmentalized expression of self-antigens in mimetic cells may explain syndromic manifestations of autoimmunity
Acknowledgments
We thank Drs. C. Evavold, R. Franklin, and S. Raychaudhuri for helpful discussions and Dr. C. Laplace for figure preparation. This work was supported by NIH grants R01AI088204 and R01DK060027 (to D.M.) and T32GM007753 (for D.A.M.).
GLOSSARY
- Agonist selection
Process by which some T-cell lineages, like Tregs and CD8αα+ T cells, are induced via a positive interaction between their TCRs and cognate peptide:MHC complexes.
- Aire
Primarily thymus-expressed TF that upregulates the expression of several thousand PTAs within mTECs. Mutations in Aire causally underlie the human autoimmune syndrome APECED (autoimmune polyendocrinopathy, candidiasis, and ectodermal dystrophy), also known as APS-1 (autoimmune polyglandular syndrome, type 1).
- Central tolerance
Process of eliminating lymphocytes that react against self-antigens; for T cells, this occurs through negative selection in the thymus. Central tolerance is augmented by peripheral tolerance, which restrains autoreactive lymphocytes that escape central tolerization.
- Cortical thymic epithelial cell (cTEC)
located within the thymic cortex; responsible for the induction of T-cell lineage commitment and positive selection into the αβ T cell lineage.
- Lineage-defining transcription factor
Protein that controls gene expression required for the differentiation, diversification, maintenance, and/or survival of a specific cellular lineage.
- Mimetic cell
mTEC that mirrors the lineage-defining TFs, chromatin-accessibility landscape, and gene-expression program of an extra-thymic cell type.
- Medullary thymic epithelial cell (mTEC)
located within the thymic medulla; responsible for the induction of PTAs, some negative selection of maturing T cells, and agonist selection of Tregs and unconventional T cells.
- Negative selection
Process by which T cells bearing autoreactive TCRs recognize their antigens on thymic antigen-presenting cells, including mTECs, and are clonally deleted from the T-cell repertoire.
- Peripheral-tissue antigen (PTA)
Protein whose expression is normally confined to extra-thymic tissues (e.g., insulin, myelin basic protein, and mucin). mTECs express thousands of PTAs.
- Positive selection
Process by which newly generated T cells are tested for the ability to interact with self-MHC molecules. T cells that successfully interact with self-MHC molecules continue to mature, while T cells lacking interaction undergo death by neglect.
- Regulatory T cell (Treg)
Foxp3+CD4+ T cell that can suppress immune responses and regulate tissue homeostasis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Abramson J et al. (2017) Thymic epithelial cells. Annu. Rev Immunol 35, 85–118 [DOI] [PubMed] [Google Scholar]
- 2.Klein L et al. (1998) CD4 T cell tolerance to human c-reative protein, an inducible serum protein, is mediated by medullary thymic epithelium. J. Exp. Med 188, 5–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kyewski B et al. (2002) Promiscuous gene expression and central T-cell tolerance: more than meets the eye. Trends Immunol 23, 364–371 [DOI] [PubMed] [Google Scholar]
- 4.Derbinski J et al. (2001) Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol 2, 1032–1039 [DOI] [PubMed] [Google Scholar]
- 5.Farr AG et al. (1998) Medullary thymic epithelium: A mosiac of epithelial "self"? J. Exp. Med 188, 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hassall AH (1846) The microscopic anatomy of the human body: in health and disease, Highley S [Google Scholar]
- 7.Remak R (1855) Untersuchungen über die Entwickelung der Wirbelthiere, Reimer G [Google Scholar]
- 8.Mayer S (1888) Zur Lehre von der Schilddrüse und Thymus bei den Amphibien. Anatomischer Anzeiger 3, 97–103 [Google Scholar]
- 9.Anderson MS et al. (2002) Projection of an immunological self shadow within the thymus by the aire protein. Science 298, 1395–1401 [DOI] [PubMed] [Google Scholar]
- 10.Aaltonen J et al. (1997) An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat Genet 17, 399–403 [DOI] [PubMed] [Google Scholar]
- 11.Nagamine K et al. (1997) Positional cloning of the APECED gene. Nat Genet 17, 393–398 [DOI] [PubMed] [Google Scholar]
- 12.Devoss J et al. (2006) Spontaneous autoimmunity prevented by thymic expression of a single self-antigen. J Exp Med 203, 2727–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gavanescu I et al. (2007) Loss of Aire-dependent thymic expression of a peripheral tissue antigen renders it a target of autoimmunity. Proc Natl. Acad Sci U S. A 104, 4583–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koh AS et al. (2008) Aire employs a histone-binding module to mediate immunological tolerance, linking chromatin regulation with organ-specific autoimmunity. Proc Natl. Acad Sci U S. A 105, 15878–15883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sansom SN et al. (2014) Population and single-cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome Res. 24, 1918–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meredith M et al. (2015) Aire controls gene expression in the thymic epithelium with ordered stochasticity. Nat Immunol 16, 942–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derbinski J et al. (2008) Promiscuous gene expression patterns in single medullary thymic epithelial cells argue for a stochastic mechanism. Proc Natl. Acad Sci U S. A 105, 657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villasenor J et al. (2008) Ectopic expression of peripheral-tissue antigens in the thymic epithelium: probabilistic, monoallelic, misinitiated. Proc Natl. Acad Sci U S. A 105, 15854–15859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinto S et al. (2013) Overlapping gene coexpression patterns in human medullary thymic epithelial cells generate self-antigen diversity. Proc Natl Acad Sci U S A 110, E3497–E3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brennecke P et al. (2015) Single-cell transcriptome analysis reveals coordinated ectopic gene-expression patterns in medullary thymic epithelial cells. Nat Immunol 16, 933–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abramson J et al. (2010) Aire's partners in the molecular control of immunological tolerance. Cell 140, 123–135 [DOI] [PubMed] [Google Scholar]
- 22.Giraud M et al. (2012) Aire unleashes stalled RNA polymerase to induce ectopic gene expression in thymic epithelial cells. Proc Natl Acad Sci U S A 109, 535–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waterfield M et al. (2014) The transcriptional regulator Aire coopts the repressive ATF7ip-MBD1 complex for the induction of immunotolerance. Nat. Immunol 15, 258–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chuprin A et al. (2015) The deacetylase Sirt1 is an essential regulator of Aire-mediated induction of central immunological tolerance. Nat Immunol 16, 737–745 [DOI] [PubMed] [Google Scholar]
- 25.Bansal K et al. (2017) The transcriptional regulator Aire binds to and activates super-enhancers. Nat Immunol 18, 263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bansal K et al. (2021) Aire regulates chromatin looping by evicting CTCF from domain boundaries and favoring accumulation of cohesin on superenhancers. Proc Natl Acad Sci U S A 118, e2110991118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farr AG et al. (2002) Organization of thymic medullary epithelial heterogeneity: implications for mechanisms of epithelial differentiation. Immunol Rev. 189, 20–27 [DOI] [PubMed] [Google Scholar]
- 28.Dooley J et al. (2005) An organized medullary epithelial structure in the normal thymus expresses molecules of respiratory epithelium and resembles the epithelial thymic rudiment of nude mice. J Immunol 175, 4331–4337 [DOI] [PubMed] [Google Scholar]
- 29.Dooley J et al. (2009) Lessons from thymic epithelial heterogeneity: FoxN1 and tissue-restricted gene expression by extrathymic, endodermally derived epithelium. J Immunol 183, 5042–5049 [DOI] [PubMed] [Google Scholar]
- 30.Gillard GO et al. (2006) Features of medullary thymic epithelium implicate postnatal development in maintaining epithelial heterogeneity and tissue-restricted antigen expression. J Immunol 176, 5815–5824 [DOI] [PubMed] [Google Scholar]
- 31.Danso-Abeam D et al. (2013) Aire mediates thymic expression and tolerance of pancreatic antigens via an unconventional transcriptional mechanism. Eur. J Immunol 43, 75–84 [DOI] [PubMed] [Google Scholar]
- 32.Bornstein C et al. (2018) Single-cell mapping of the thymic stroma identifies IL-25-producing tuft epithelial cells. Nature 559, 622–626 [DOI] [PubMed] [Google Scholar]
- 33.Miller CN et al. (2018) Thymic tuft cells promote an IL-4-enriched medulla and shape thymocyte development. Nature 559, 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dhalla F et al. (2020) Biologically indeterminate yet ordered promiscuous gene expression in single medullary thymic epithelial cells. EMBO J 39, e101828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baran-Gale J et al. (2020) Ageing compromises mouse thymus function and remodels epithelial cell differentiation. eLife 9, e56221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park JE et al. (2020) A cell atlas of human thymic development defines T cell repertoire formation. Science 367, eaay3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bautista JL et al. (2021) Single-cell transcriptional profiling of human thymic stroma uncovers novel cellular heterogeneity in the thymic medulla. Nat Commun 12, 1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michelson DA et al. (2022) Thymic epithelial cells co-opt lineage-defining transcription factors to eliminate autoreactive T cells. Cell 185, 2542–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Givony T et al. (2022) Thymic microfold and endocrine cells regulate thymus homeostasis and self-tolerance. Research Square, https://assets.researchsquare.com/files/rs-1837610/v1_covered.pdf?c=1657825917 [Google Scholar]
- 40.Wells KL et al. (2020) Combined transient ablation and single-cell RNA-sequencing reveals the development of medullary thymic epithelial cells. eLife 9, e60188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watanabe N et al. (2005) Hassall's corpuscles instruct dendritic cells to induce CD4+CD25+ regulatory T cells in human thymus. Nature. 436, 1181–1185 [DOI] [PubMed] [Google Scholar]
- 42.Koh AS et al. (2018) Rapid chromatin repression by Aire provides precise control of immune tolerance. Nat Immunol 19, 162–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Org T et al. (2009) AIRE activated tissue specific genes have histone modifications associated with inactive chromatin. Hum. Mol. Genet 18, 4699–4710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koh AS et al. (2010) Global relevance of Aire binding to hypomethylated lysine-4 of histone-3. Proc Natl Acad Sci U S A 107, 13016–13021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaiser C et al. (2022) AIRE in context: Leveraging chromatin plasticity to trigger ectopic gene expression. Immunol Rev 305, 59–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guerau-de-Arellano M et al. (2009) Neonatal tolerance revisited: a perinatal window for Aire control of autoimmunity. J Exp. Med 206, 1245–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murata S et al. (2007) Regulation of CD8+ T cell development by thymus-specific proteasomes. Science 316, 1349–1353 [DOI] [PubMed] [Google Scholar]
- 48.Bell GI et al. (1984) A polymorphic locus near the human insulin gene is associated with insulin-dependent diabetes mellitus. Diabetes 33, 176–183 [DOI] [PubMed] [Google Scholar]
- 49.Vafiadis P et al. (1997) Insulin expression in human thymus is modulated by INS VNTR alleles at the IDDM2 locus. Nat. Genet 15, 289–292 [DOI] [PubMed] [Google Scholar]
- 50.Pugliese A et al. (1997) The insulin gene is transcribed in the human thymus and transcription levels correlated with allelic variation at the INS VNTR-IDDM2 susceptibility locus for type 1 diabetes. Nat. Genet 15, 293–297 [DOI] [PubMed] [Google Scholar]
- 51.Barrett JC et al. (2009) Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet 41, 1330–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu X et al. (2010) Genome-wide meta-analyses identify three loci associated with primary biliary cirrhosis. Nat Genet 42, 658–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.International Multiple Sclerosis Genetics Consortium et al. (2011) Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476, 214–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmidt D et al. (1997) A mechanism for the major histocompatibility complex-linked resistance to autoimmunity. J. Exp. Med 186, 1059–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taneja V et al. (2008) Delineating the role of the HLA-DR4 "shared epitope" in susceptibility versus resistance to develop arthritis. J Immunol 181, 2869–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ooi JD et al. (2017) Dominant protection from HLA-linked autoimmunity by antigen-specific regulatory T cells. Nature 545, 243–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ishigaki K et al. (2022) HLA autoimmune risk alleles restrict the hypervariable region of T cell receptors. Nat Genet 54, 393–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gunther T et al. (2000) Genetic ablation of parathyroid glands reveals another source of parathyroid hormone. Nature 406, 199–203 [DOI] [PubMed] [Google Scholar]
- 59.Liu Z et al. (2010) Thymus-associated parathyroid hormone has two cellular origins with distinct endocrine and immunological functions. PLoS Genet. 6, e1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Panneck AR et al. (2014) Cholinergic epithelial cell with chemosensory traits in murine thymic medulla. Cell Tissue Res. 358, 737–748 [DOI] [PMC free article] [PubMed] [Google Scholar]