Figure 3: Cell Painting profiles identify compounds impacting PPARGC1A (PGC-1α) overexpression phenotypes.
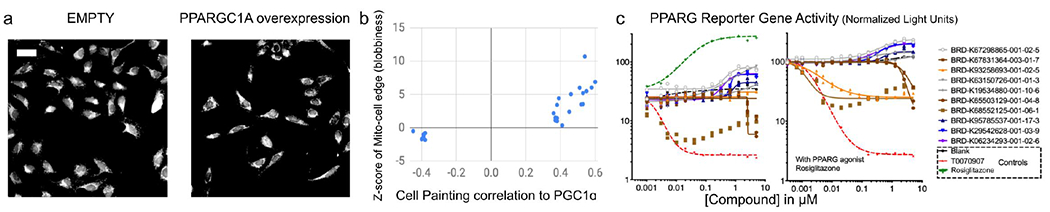
a) Cell Painting images for PPARGC1A (PGC-1α) overexpression compared to negative control (EMPTY, same field as in Figure 1a). Scale bar = 60 ϋm. b) Compounds with high or low correlations of their Cell Painting profiles to PGC-1α overexpression were chosen for further study (hence all samples are below ~−0.35 or above ~0.35 on the X axis, which shows the Pearson correlation between the single, consensus gene profile and the single, consensus compound profile. The consensus profiles are created using median across 5 replicates for gene overexpressions, eight replicates for bioactive compounds, and four replicates for diversity-oriented synthesis (DOS) compounds). Correlation to PGC-1α overexpression is dominated by one feature, the standard deviation of the MitoTracker staining intensity at the edge of the cell, which we term blobbiness (displayed on the Y axis as a z-score with respect to the negative controls and is calculated based on eight replicates for bioactive compounds, and four replicates for DOS compounds). c) PPARG reporter gene assay dose-response curves in the absence (left) or presence (right) of added PPARG agonist, Rosiglitazone. The ten most active compounds are shown and reported as normalized light units; data representative of two experiments; two replicate data points are shown for each BRD compound but sometimes overlap. Line of best fit generated using GraphPad PRISM using variable slope four parameters fit of log(inhibitor) vs. response. Compounds highlighted in blue/purple are structurally related pyrazolo-pyrimidines.
