Figure 5. Effects of Runx2 gain of function on acquisition of chromatin accessibility in fibroblasts.
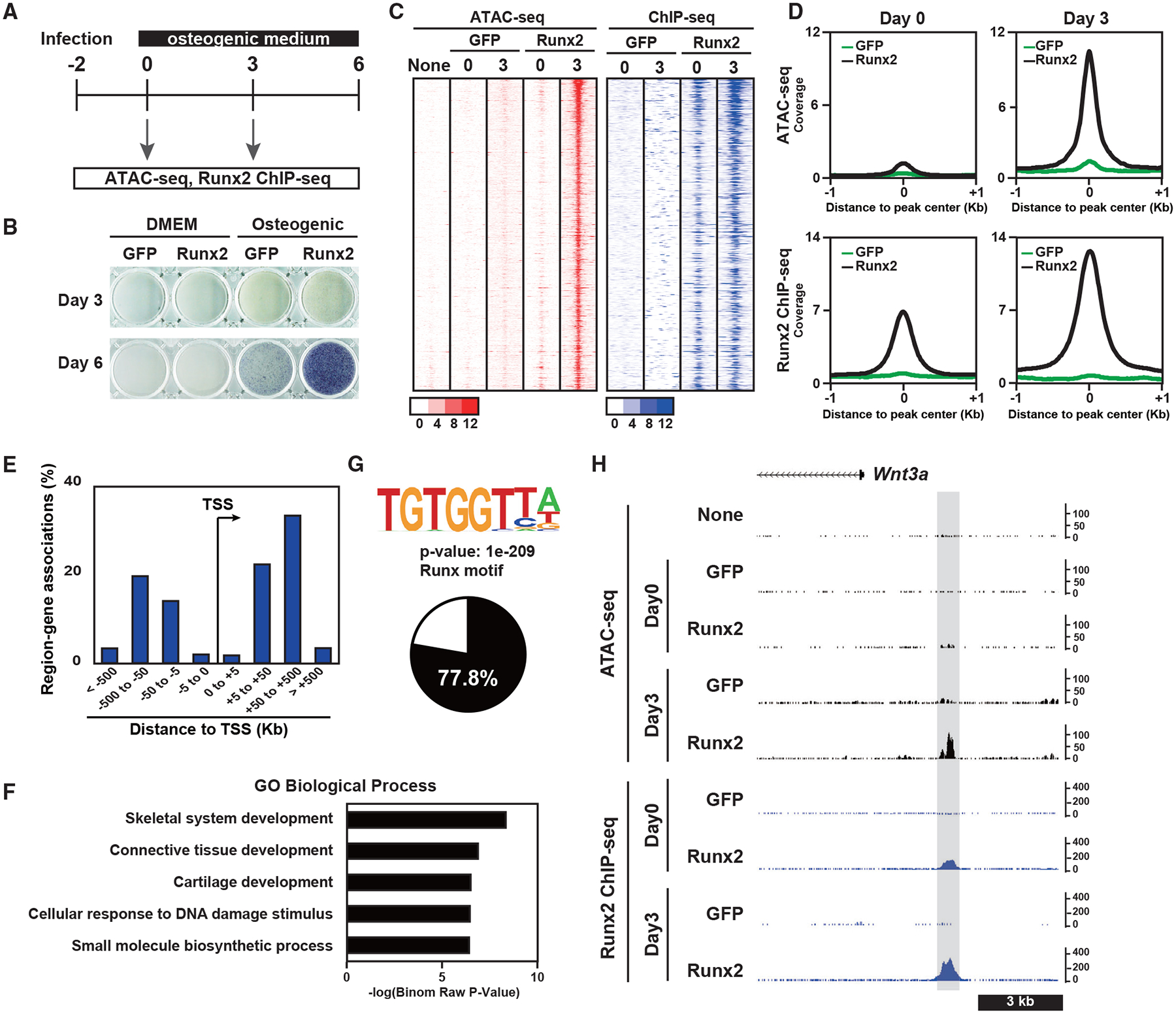
(A) Schematic of the experiment.
(B) ALP staining of NIH 3T3 fibroblasts overexpressing GFP or Runx2-BioFL. Cells were cultured with DMEM or an osteogenic medium.
(C) Heatmap showing the signal intensities of ATAC-seq and Runx2 ChIP-seq of NIH 3T3 cells overexpressing GFP or Runx2-BioFL for the indicated culture periods. We selected 731 genomic regions representing the pioneer action of Runx2. Cells were cultured in an osteogenic medium.
(D) Histogram showing the average of normalized signals in ATAC-seq (top) and Runx2 ChIP-seq (bottom) on day 0 (left) and day 3 (right) in a 2-kb window from the center of the peaks. The values were normalized from biological duplicates.
(E) Genome-wide distribution of Runx2-responsive chromatin-accessible regions relative to TSSs.
(F) GREAT GO annotations of genomic regions representing the pioneer action of Runx2.
(G) Motif logo of de novo motif analysis of 731 genomic regions representing the pioneer action of Runx2 (top) and a pie chart showing the percentage of peaks containing the Runx consensus motif in these genomic regions (bottom).
(H) CisGenome browser screenshot of the flanking region of Wnt3a showing chromatin accessibility and Runx2-DNA binding in NIH 3T3 cells overexpressing GFP or Runx2-BioFL. A genomic region representing the pioneer action of Runx2 is highlighted in gray.
