Figure 6. Screening for putative enhancers targeted by Runx2 in Obs.
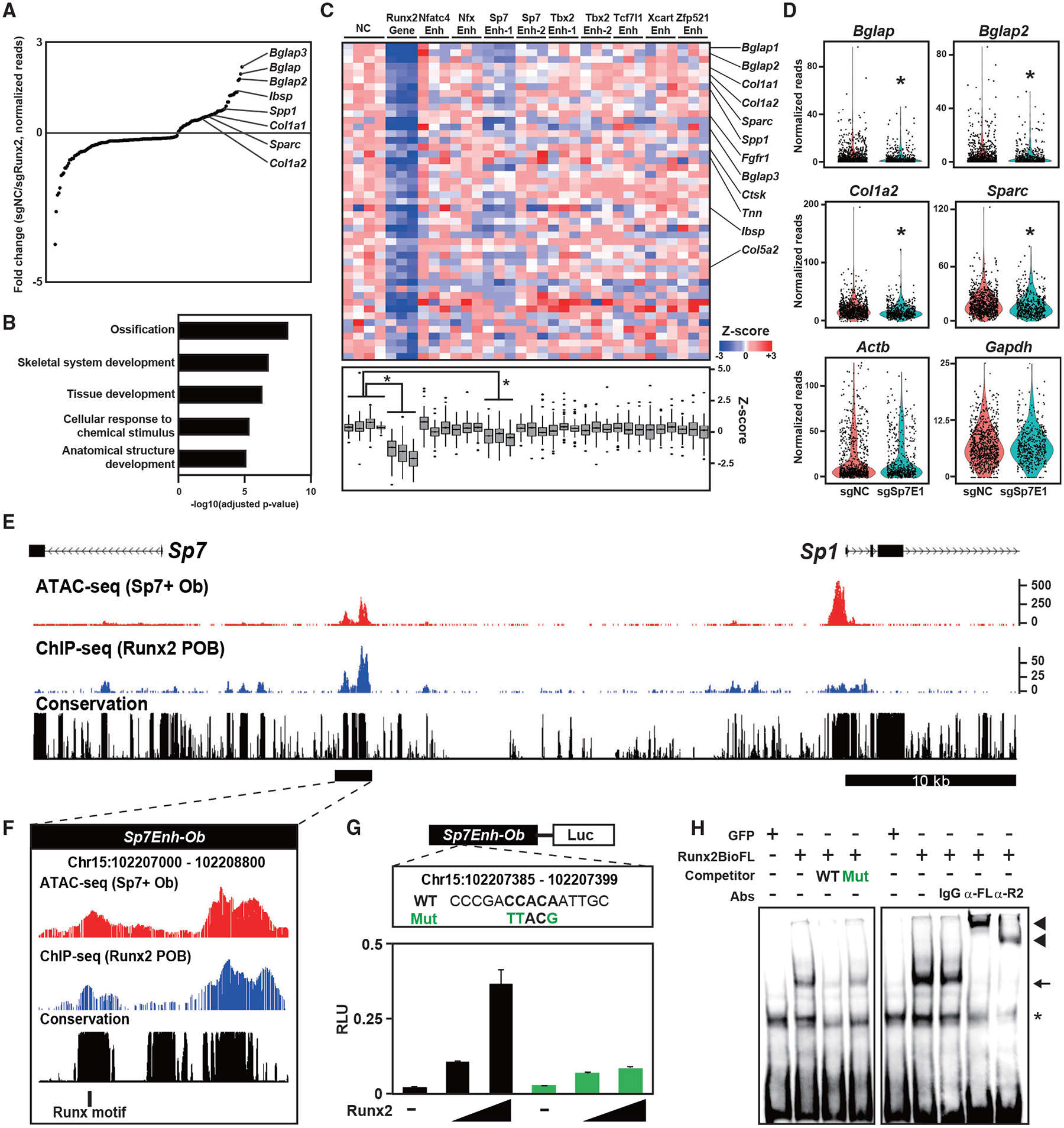
(A) Dot plots showing genes with significantly altered expression induced by sgRunx2 expression. Ob marker genes are highlighted. Normalized values in cells expressing sgRNAs for negative control sequences (sgNC, n = 820) and those for Runx2 (sgRunx2, n = 312) were used.
(B) g:Profiler GO annotations of biological processes in genes with significantly decreased expression induced by sgRunx2 expression.
(C) Heatmap showing Z scores representing relative gene expression in cells expressing the indicated sgRNAs (top) and boxplot of Z scores (bottom). Genes with significantly decreased expression induced by sgRunx2 were selected. Ob-related genes are indicated on the right. *p < 0.05 versus cells expressing sgNC in the one factor-associated site (Tukey-HSD analysis). Normalized values in cells expressing each gRNA (n = 87–368) were used.
(D) Violin plot showing normalized reads of the indicated genes in cells expressing sgNC or Sp7 Enhancer 1 (sgSp7E1). Each dot represents a cell. *p < 5.0 × 10−4 versus cells expressing sgNC. Normalized values from sgNC (n = 820) and sgSp7E1 (n = 498) were used.
(E and F) CisGenome browser screenshot of the Sp7Enh-Ob region showing chromatin accessibility and Runx2-DNA binding in Obs. The region indicated by the black bar in (E) is enlarged in (F), indicating a Runx motif targeted by sgSp7E1.
(G) Schematic of the luciferase reporter driven by Sp7Enh-Ob (top) and luciferase reporter assay in 293T cells transfected with WT and mutated (Mut) reporter constructs together with different amounts of Runx2-expressing plasmids (bottom). WT and Mut DNA sequences of the Runx motif targeted by sgSp7E1 (top) are shown. Data are presented as the means ± SD of triplicate experiments.
(H) EMSA of Runx2-BioFL using a probe containing the Runx motif targeted by sgSp7E1. WT and Mut probes, whose sequences are shown in (G), were used for the competitor assay. Anti-FL (α-FL) and anti-Runx2 antibodies (α-R2) and an immunoglobulin G (IgG) negative control were used for the supershift assay. Arrow, RUNX2-DNA complex; arrowhead, RUNX2-DNA-antibody complex; asterisk, non-specific signal.
See also Figures S6 and S7 and Table S6.
