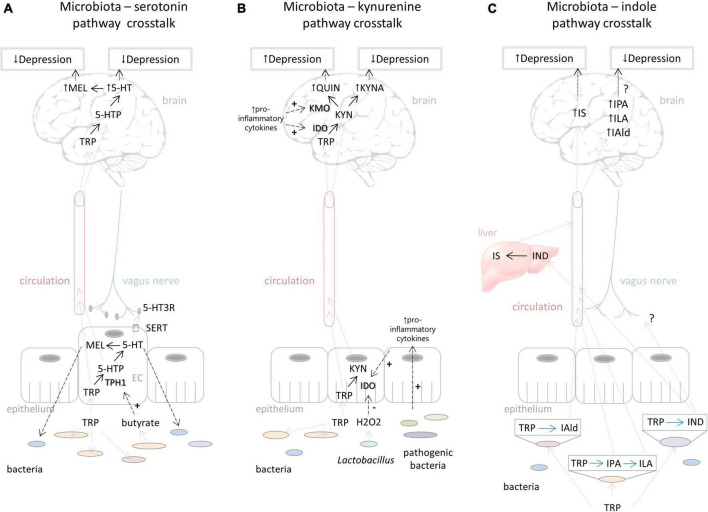
FIGURE 2.
Putative ways of gut microbiota – host interactions in the context of the tryptophan metabolism influencing depressive behavior. (A) Effects of microbiota on serotonin pathway include regulation of tryptophan availability which directly influences serotonin and probably melatonin synthesis in the brain affecting depressed mood. Microbiota can stimulate TPH1 activity by its metabolites (e.g., butyrate). Serotonin produced in the gut could have an impact on brain functioning via stimulation of the vagus nerve. On the other hand, host serotonin and melatonin synthesis in the gut can affect the commensal bacterial composition, contributing individual’s susceptibility to depression. (B) Bacterial effects on the kynurenine pathway include down-regulation of IDO activity by H2O2 produced by Lactobacillus. The proliferation of pathogenic bacteria that stimulates pro-inflammatory cytokine production stimulates IDO activity and QUIN branch of the kynurenine pathway. Increased QUIN is associated with depression. On the other hand, increased KYNA levels could be related to reduction of depressive symptoms. (C) Depending on the specific catalytic enzymes that different bacterial species harbor, tryptophan can be degraded into various indolic compounds. They are absorbed by the host into the circulation and can be further metabolized in lever; for example, indole is metabolized to IS, which could, in high concentrations, lead to depressive behavior. Various indolic compounds (e.g., IPA, ILA, IAld) were shown to have neuroprotective effects, but their effects on depression are still unexplored. For more details, see explanations in the text. Black dashed arrows represent directions of effects (“+” - represents stimulation, “-” - represents inhibition). Gray dashed arrows represent directions of movement of metabolites. Black dotted arrows represent effects on depression. Black full arrows represent host pathways. Blue full arrows represent bacterial pathways. Question marks represent effects that are not still well explored. TRP, tryptophan; TPH, tryptophan hydroxylase; 5-HTP, 5-hydroxytryptophan; 5-HT, 5-hydroxytryptamine (serotonin); MEL, melatonin; SERT, serotonin transporter; EC, enterochromaffin cells; IDO, indolamine 2,3-dioxygenase; TDO, tryptophan 2,3-dioxygenase; KYN, kynurenine; KYNA, kynurenine acid; KMO, kynurenine 3-monooxygenase; QUIN, quinolinic acid; IAld, indole-3-aldehyde; IPA, indole-3-propionic acid; ILA, indole-3-lactic acid; IND, indole; IS, indoxil-3-sulfate.