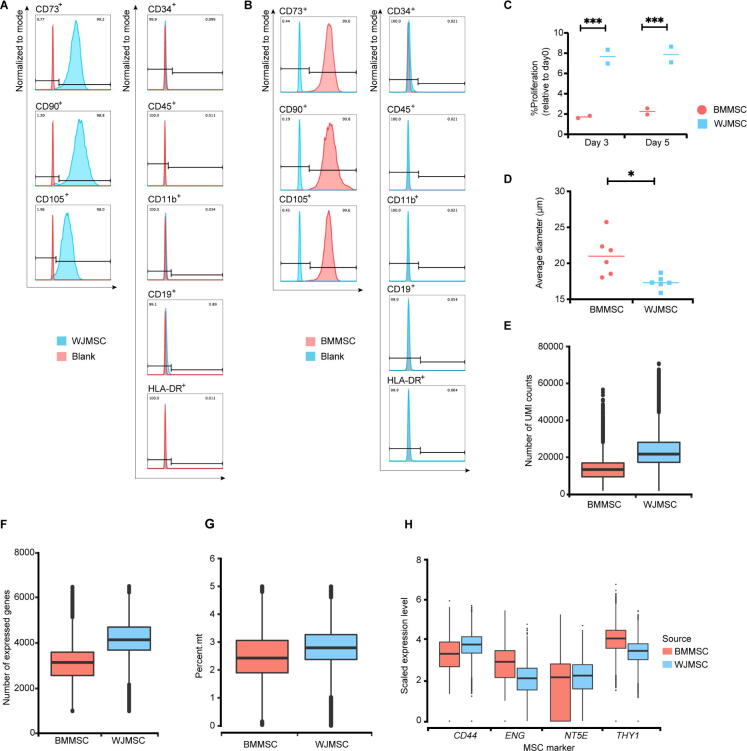
Supplementary figure S1.
Characteristics of MSCs and quality control of scRNA-seq data. A. A representative flow diagram of the expression of positive markers (CD73, CD90, and CD105) and negative markers (CD34, CD45, CD11b, CD19, and HLA-DR) in WJMSCs. The proportion of cells with positive or negative expression is listed in the upper right or upper left corner. B. A representative flow diagram of the expression of positive markers (CD73, CD90 and CD105) and negative markers (CD34, CD45, CD11b, CD19, and HLA-DR) in BMMSCs. The proportion of cells with positive expression or negative expression is listed in the upper right or upper left corner. C. Histogram showing the proliferation ability as evaluated by the ratios of the cell counts on day 3 and day 5 relative to that on day 0. (n = 2, the data are expressed as the mean ± SD. ***, P < 0.005; analyzed by t-test.) D. Histogram showing the average diameters as analyzed with a Countstar instrument. (n = 6, the data are expressed as the mean ± SD. *, P < 0.05; analyzed by t-test.). E.–G. Box plot showing the number of UMI counts (E), expressed genes (F), and the percentage of mitochondrial genes (G) from single-cell transcriptomic data in WJMSCs and BMMSCs. H. Histogram showing the expression patterns of positive marker genes (CD73/NT5E, CD90/THY1, CD105/ENG, and CD44). The data are expressed as the mean ± SD. Percent.mt, the percentage of mitochondrial genes; UMI, unique molecular identifier; scRNA-seq, single cell RNA-sequencing.