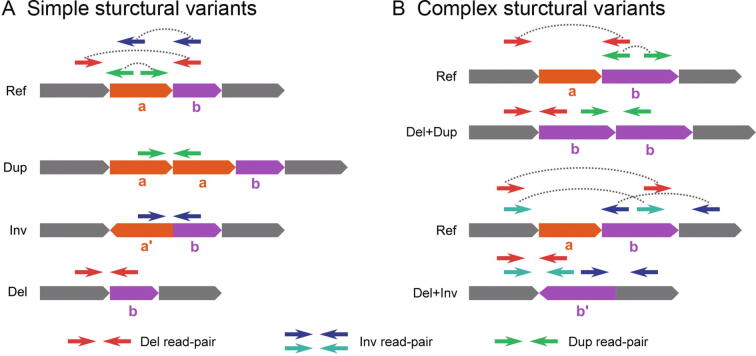
Figure 1.
Explanation of simple and complexSValignment models derived from abnormal read-pairs
A. Three common simple SVs and their corresponding abnormal read-pair alignments on the reference genome. B. The alignment signature of two CSVs. Each involves two types of signatures that can be matched by a simple SV alignment model. SV, structural variant; CSV, complex structural variant; Ref, reference; Dup, duplication; Inv, inversion; Del, deletion.