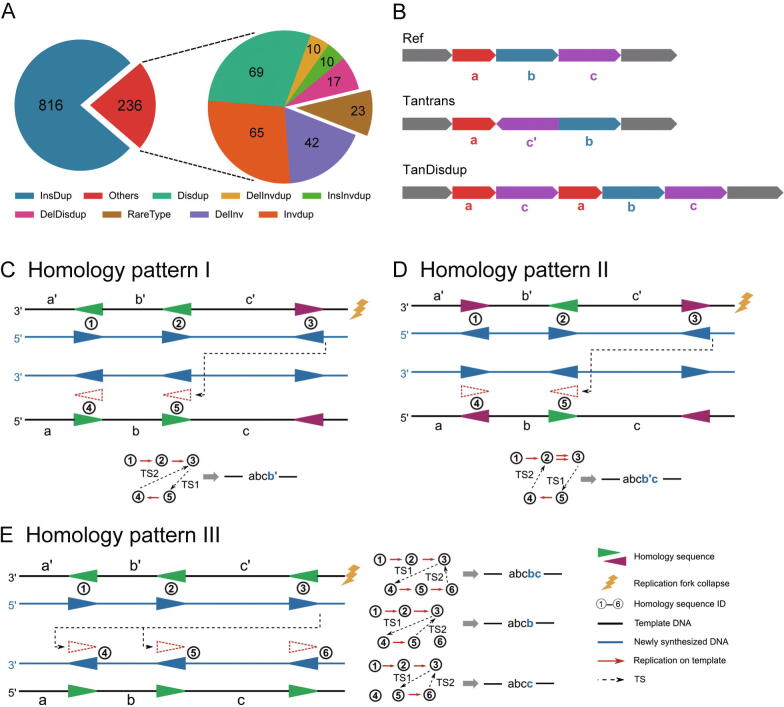
Figure 6.
Overview of Mako’s CSV discoveries from three healthy samples and proposed CSV formation mechanisms
A. Summary of discovered CSV types. These types are reconstructed by PacBio HiFi reads, where a type with less than 10 events is summarized as RareType. B. Diagrams of two novel and rare CSV types discovered by Mako. In particular, Mako finds three Tantrans events and only one TanDisdup event. C.–E. Different replication diagrams explaining the impact of homology pattern for MMBIR-produced CSVs. In these diagrams, sequence abc has been replicated before the replication fork collapse (flash symbol). The single-strand DNA at the DNA DSB starts searching for homology sequence (purple and green triangles) to repair. The a forementioned procedure is explicitly explained as a replication graph, where nodes are homology sequences and edges keep track of TS (dotted arrow lines) as well as the normal replication at different strands (red lines). If there are two red lines between two nodes, the sequence between these two nodes will be replicate twice, as shown in (D). InsDup, insertion associated with duplication; Disdup, dispersed duplication; Invdup, inverted duplication; DelInvdup, deletion associated with inverted duplication; InsInvdup, insertion associated with inverted duplication; DelDisdup, deletion associated with dispersed duplication; DelInv, deletion associated with inversion; Tantrans, adjacent segment swap; TanDisdup, tandem dispersed duplication; MMBIR, microhomology-mediated break-induced replication; DSB, double-strand break; TS, template switch.