Figure 3.
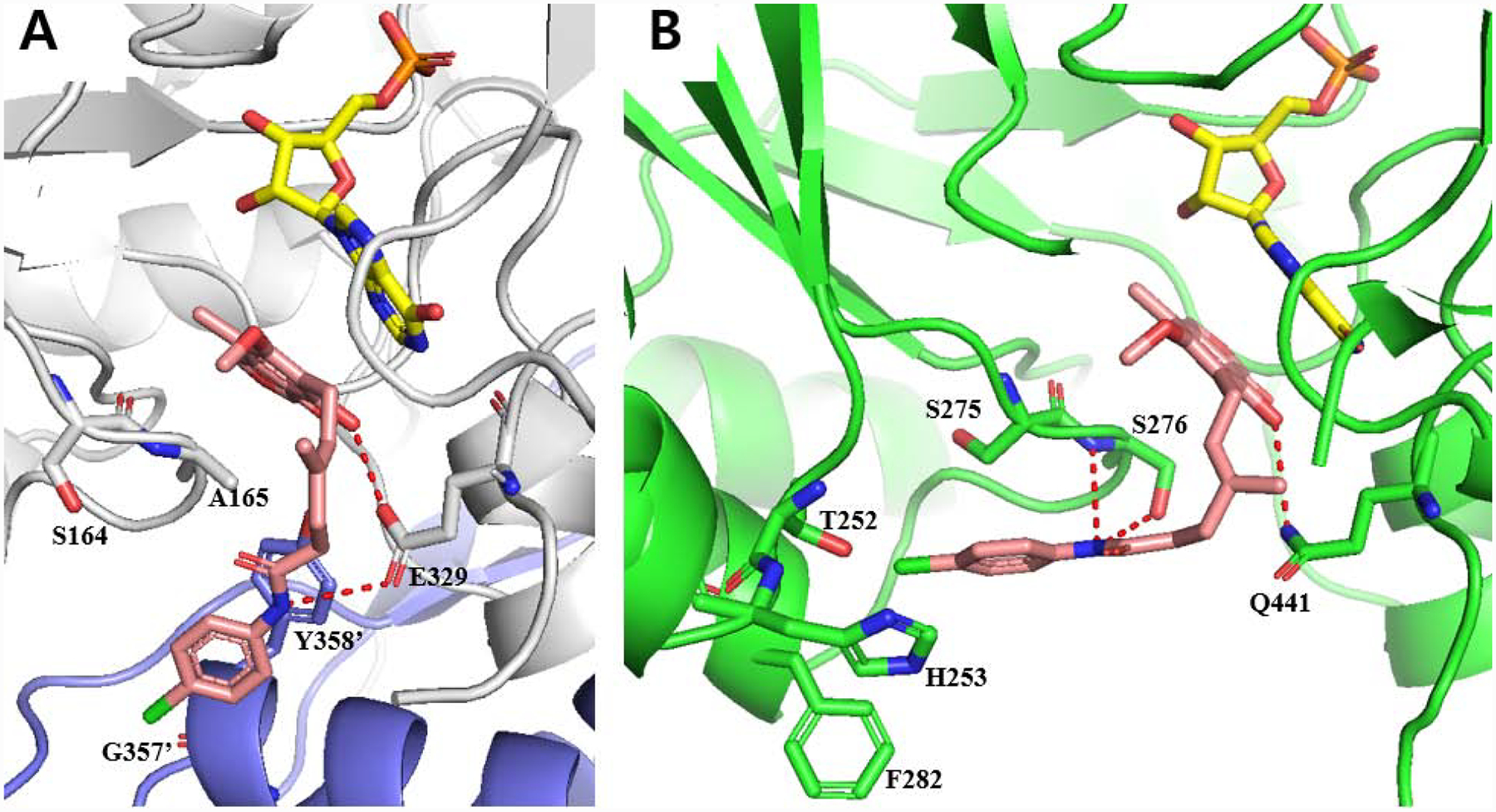
(A) Docked model of 12 (pink) with CpIMPDH (gray, PDB: 3KHJ) and IMP (yellow). Adjacent monomer protein is shown as purple and residue numbers are differentiate by prime (‘). Hydrogen bonds are shown as red dashes (< 3.0 Å). Docking score of 12 with CpIMPDH•IMP was −9.86. (B) Docked structure of 12 (pink) with hamster IMPDH2 (green, PDB: 1JR1) and IMP (yellow), which has only eight non-binding site amino acid differences compared to HsIMPDH243. Hydrogen bonds are shown as red dashes (< 3.5 Å). Docking score of 12 with hamster IMPDH2•IMP was −8.83. Water, K+ and other protein subunits were deleted for docking and presentation for clarity.
