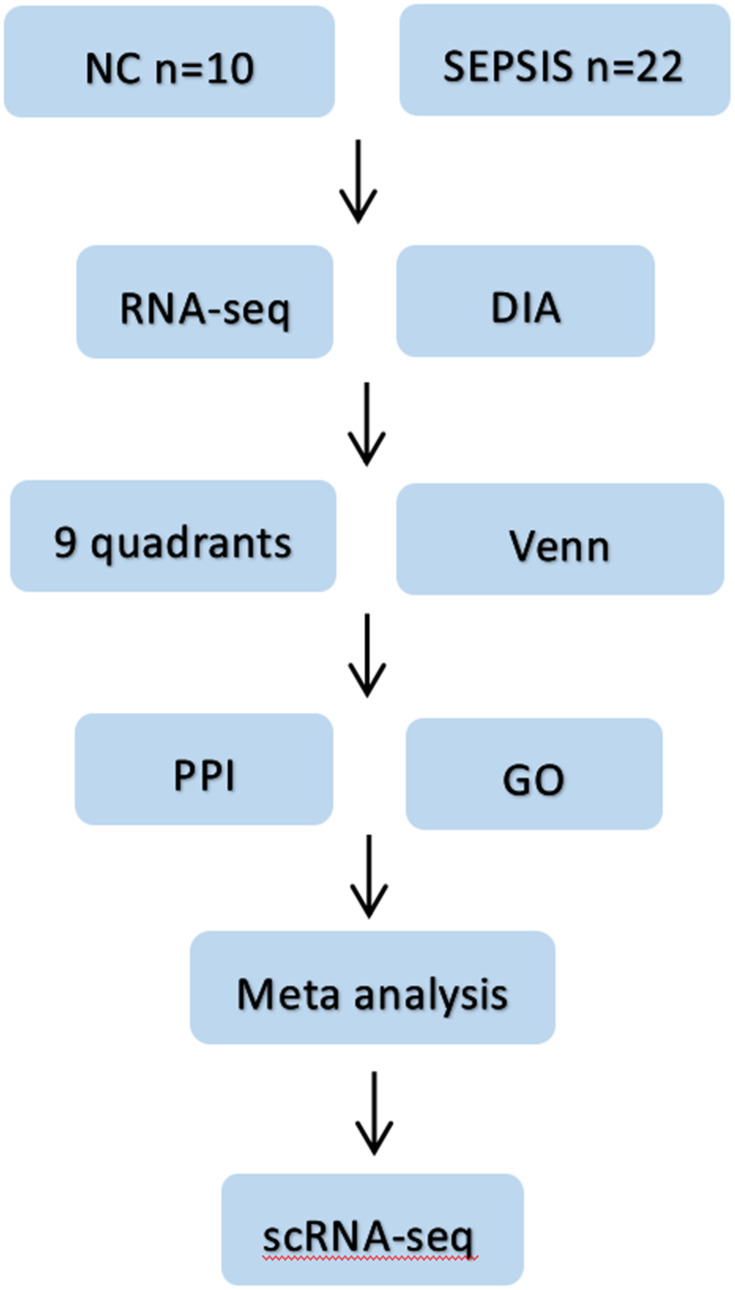
Figure 1.
Flow chart of the study. Firstly, blood samples from sepsis patients and normal subjects were collected, then differential proteins and differential genes were screened by DIA protein profile and RNA sequencing combined with bioinformatics methods; then proteomics and genomics data were jointly analyzed; later, intersecting targets were taken for PPI and GO analysis, and four potential core genes were finally clarified by meta-analysis. Finally, scRNA-seq was used to clarify the cell lines localization of the target genes.