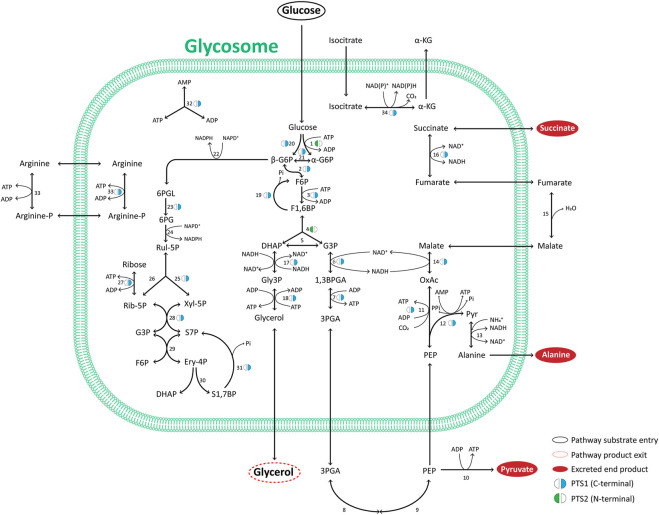
FIGURE 2.
Major pathways of glycosomal carbon metabolism in TriTryps species. Indicated are enzymes of glycolysis, its auxiliary branches, gluconeogenesis, glycerol metabolism and the pentose-phosphate pathway. Note that not all enzymes indicated are present in each of the TriTryps species or expressed in each of their life-cycle stages. Enzymes: 1. Hexokinase; 2. Glucose-6-phosphate isomerase; 3. Phosphofructokinase; 4. Aldolase; 5. Triose-phosphate isomerase; 6. Glyceraldehyde-3-phosphate dehydrogenase; 7. Phosphoglycerate kinase; 8. Phosphoglycerate mutase; 9. Enolase; 10. Pyruvate kinase; 11. Phosphoenolpyruvate carboxykinase; 12. Pyruvate phosphate dikinase; 13. Alanine dehydrogenase; 14. Malate dehydrogenase; 15. Fumarate hydratase; 16. NADH-dependent fumarate reductase; 17. Glycerol-3-phosphate dehydrogenase; 18. Glycerol kinase; 19. Fructose-1,6-bisphosphatase; 20. Glucokinase; 21. d-Hexose-6-Phosphate-1-epimerase; 22. Glucose-6-phosphate dehydrogenase; 23. 6-Phosphogluconolactonase; 24; 6-Phosphogluconate dehydrogenase; 25. Ribulose-5-phosphate epimerase; 26. Ribulose-5-phosphate isomerase; 27. Ribokinase; 28. Transketolase; 29. Transaldolase; 30. Sedoheptulose-1,7-phosphate transaldolase; 31. Sedoheptulose-1,7-bisphosphatase; 32. Adenylate kinase; 33. Arginine kinase; 34. Isocitrate dehydrogenase. The presence of a PTS1 or 2, the carbon sources used, and the excreted end products are indicated.