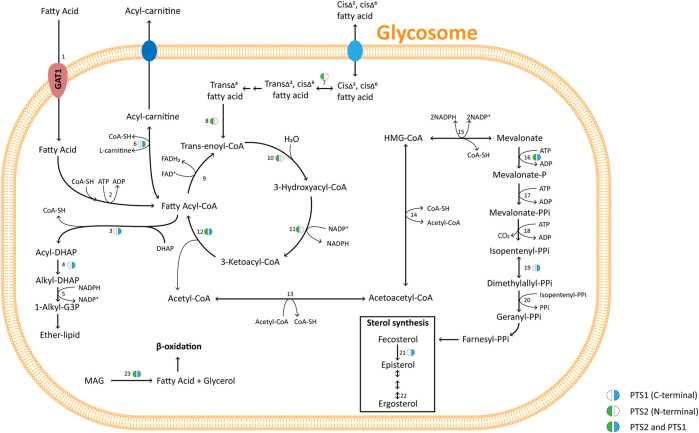
FIGURE 5.
Pathways of glycosomal lipid metabolism in TriTryps species. Pathways of ether-lipid synthesis (enzymes 3 – 5), fatty-acid β-oxidation (9 – 12), and sterol synthesis (16 – 23) are represented. Note that not all enzymes indicated are present in each of the TriTryps species or expressed in each of their life-cycle stages, and for simplicity not all reactions are represented. Enzymes/transporter: 1. Glycosomal ABC transporter (GAT1); 2. Fatty-acyl CoA synthetase; 3. DHAP acyltransferase; 4. Alkyl-dihydroxyacetone phosphate synthase; 5: 1-Alkyl G3P:NADP+ oxidoreductase; 6. Carnitine O-palmitoyltransferase; 7. 3,2-Trans-enoyl-CoA isomerase; 8. Enoyl-CoA isomerase; 9. Acyl-CoA dehydrogenase; 10, 11, 12. Enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase (trifunctional enzyme); 13. Acetyl-CoA acetyltransferase 14. 3-Hydroxy-3-methylglutaryl CoA synthase; 15. 3-Hydroxy-3-methylglutaryl-CoA reductase; 16. Mevalonate kinase; 17. Phosphomevalonate kinase 18. Mevalonate diphosphate decarboxylase 19. Isopentenyl-diphosphate δ-isomerase; 20. Farnesyl diphosphate synthase 21. C-8 sterol isomerase; 22. Sterol C-24 reductase; 23. Monoacylglyceride lipase. MAG: monoacylglycerol. Enzymes 13, 14, 15, 17, 18 and 20 have been detected experimentally with a PTS motif in mammalian peroxisomes (references cited in Acosta et al., 2019).