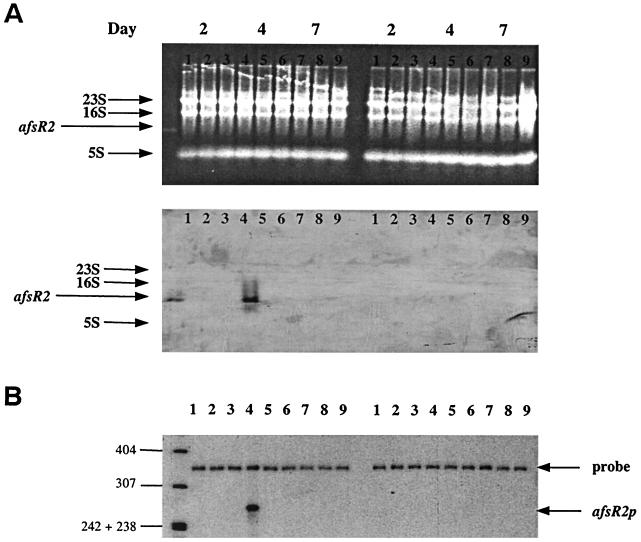
FIG. 4.
(Top) Northern blot analysis of total RNA isolated from S. lividans TK21 (lanes 1, 4, and 7), SL94 (lanes 2, 5, and 8), and SL41 (lanes 3, 6, and 9) grown on minimal-medium plates containing either glycerol (left) or glucose (right) as the sole carbon source. Total RNAs from the three strains were isolated at day 2 (substrate mycelia; lanes 1, 2, and 3), day 4 (aerial mycelia; lanes 4, 5, and 6), and day 7 (spores; lanes 7, 8, and 9). The top panel, which indicates equal loading of samples into lanes, shows the ethidium bromide-stained gel, indicating the locations of rRNA. The location of purified afsR2 mRNA isolated by prehybridization with afsR2 probe DNA which was the same as that used for S1 mapping (lane at extreme left). The lower panel shows the Northern blot. (Bottom) S1 nuclease protection analysis of transcription of afsR2 during development occurring in surface-grown cultures of S. lividans TK21 (lanes 1, 4, and 7), SL94 (lanes 2, 5, and 8), and SL41 (lanes 3, 6, and 9). RNA was isolated from cells grown on cellophane-covered minimal-medium plates containing either glycerol (left) or glucose (right) as the sole carbon source for 2 days (substrate mycelia; lanes 1, 2, and 3), 4 days (aerial mycelia; lanes 4, 5, and 6), and 7 days (spores; lanes 7, 8, and 9). The uniquely 5′-end-labeled probes were prepared as described in Materials and Methods. A protected 282-bp fragment representing afsR2 transcripts detected only in S. lividans TK21 (afsR2+) grown on glycerol minimal-medium plates for 4 days (lane 4). The position of DNA size marker bands is shown to the left of the figure.