Abstract
This study aimed at investigating the nature of SARS-CoV-2-specific immunity in patients with mild COVID-19 and sought to identify parameters most relevant for the generation of neutralizing antibody responses in convalescent COVID-19 patients. In the majority of the examined patients a cellular as well as humoral immune response directed to SARS-CoV-2 was detected. The finding of an anti-SARS-CoV-2-reactive cellular immune response in healthy individuals suggests a pre-existing immunity to various common cold HCoVs which share close homology with SARS-CoV-2. The humoral immunity to the S protein of SARS-CoV-2 detected in convalescent COVID-19 patients correlates with the presence of SARS-CoV-2-reactive CD4+ T cells expressing Th1 cytokines. Remarkably, an inverse correlation of SARS-CoV-2 S protein-specific IgGs with HCoV-NL63 and HCoV-229E S1 protein-specific IgGs suggests that pre-existing immunity to Alphacoronaviruses might have had an inhibitory imprint on the immune response to SARS-CoV-2-infection in the examined patients with mild COVID-19.
Keywords: SARS-CoV-2, COVID-19, SARS-CoV-2-reactive T-cell immunity, Humoral immunity, Common cold human Coronavirus, Alpha- and Betacoronavirus
Abbreviations
- AbR
Antibody ratio
- CD
Cluster of differentiation
- COVID-19
coronavirus disease 2019
- ELISA
Enzyme-linked immunosorbent assay
- EMA
Ethidium monoazide
- ICS
intracellular cytokine staining
- HI
healthy individual
- HSA
human serum albumin
- HCoV
Human Coronavirus
- IFN-γ
Interferon-gamma
- IL-2
Interleukin 2
- LOD
limit of detection
- MABA
multiplex antibody bead array
- MERS
Middle East Respiratory Syndrome
- MFI
Mean Fluorescence Intensity
- PBMCs
peripheral blood mononuclear cells
- PRNT
plaque reduction neutralization test
- RBD
Receptor binding domain
- SARS
Severe Acute Respiratory Syndrome
- Th1
T helper cell type 1
- TNF-α
Tumor necrosis factor α.
1. Introduction
Infection with the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) causes the coronavirus disease 2019 (COVID-19) in approximately 5–15% of infected individuals. SARS-CoV-2 is a large RNA virus belonging to the Betacoronavirus genus within the family of Coronaviridae [1] infecting a wide range of hosts including man using angiotensin-converting enzyme 2 (ACE2) receptor as the dominant mechanism of cell entry [2]. While SARS-CoV and Middle East Respiratory Syndrome (MERS)-CoV have caused limited epidemics with severe pneumonia, other members of the human CoV-family, namely HCoV-OC43, HCoV-HKU1, HCoV-229E and HCoV-NL63, are endemically transmitted causing common cold with rare fatal infections of the upper and lower respiratory tract [3].
Infections with HCoV result in specific cellular and humoral immunity that can be detected several years after infection. While HCoV-specific T-cell memory remains stable and can be detected more than 11 years after SARS-CoV infection ([4, 5]), SARS-CoV- and HCoV-229E-specific antibodies titers have been shown to decline and fall below the detection limit within few years in a significant proportion of infected individuals [6], [7], [8], [9]. However, this is not surprising and a common phenomenon for primary immune responses leading to fast clearance of the pathogen.
Among members of the HCoVs many proteins, particularly the ORF1 region, are highly conserved and hence show a high homology among many HCoVs [10]. Therefore, cross-reactivity of HCoV-specific T cells with SARS-CoV-2 antigens present in individuals might contribute to the susceptibility and severity of SARS-CoV-2 infection and COVID-19 [5]. Indeed, SARS-CoV-specific T-cells responses were detected in a significant proportion of SARS-Co-2 unexposed individuals [5].
Significant reduced number of particularly CD8+ T cells found in peripheral blood of patients with moderate and severe COVID-19 cases were reported [11], [12], [13]. In contrast, the vast majority of convalescent patients or patients with mild symptoms show normal or slightly increased T cell counts [14], [15], [16]. The magnitude of T-cell lymphopenia correlates with the severity and mortality of COVID-19, thereby demonstrating the pivotal role of which T cells play for the course of the disease ([12, 13, 15]).
In convalescent COVID-19 patients the SARS-CoV-specific T-cell immunity is dominated by CD8+ T-cells directed to various structural viral proteins including M, N, S as well as ORF3, whereas CD4+ T-cell immunity was found to be mainly confined to the S protein [17]. However, T-cell specificity for SARS-CoV-2 declined and in only one-third of patients SARS-CoV-2 T-cell immunity restricted to the viral N protein was detected post recovery [18]. Another study demonstrated that SARS-CoV-2-specific CD4+ and CD8+ T-cell responses are predominantly directed to the viral S protein epitopes in COVID-19 patients with moderate to severe acute respiratory distress symptoms approximately two weeks after admission to an intensive care unit. Since durable high-affinity antibody responses depend on CD4+ T-cell help, key to the understanding of the generation of SARS-CoV-2 neutralizing antibodies and severity of the disease is the CD4+ T-cell immunity directed to the receptor binding domain (RBD) of SARS-CoV-2. The extent of humoral cross-reactivity between Alpha- and Betacoronaviruses has been analysed in natural and experimental infections studies. Antibody responses directed to common cold HCoV N proteins indicate that cross-reactivity is limited within the Alpha-HCoVs (HCov229E and HCoVNL63) and Beta-HCoVs (HCoVOK43 and HCoVHUK1), but does not occur between Alpha-HCoVs and Beta-HCoVs [19], [20], [21], [22]. Moreover, no or little humoral cross-reactivity has been observed between common cold HCoVs, SARS-CoV and MERS [23], [24], [25], [26], [27].
2. Material and methods
2.1. Convalescent COVID-19 patients and healthy individuals before 2019
Heparinized whole blood samples from 15 healthy individuals (HI) collected before 2019 (age: 45.7 ± 12.8 (mean and SD)) and 18 SARS-CoV-2 convalescent COVID-19 patients (age: 35.6 ± 10.3 (mean and SD)) were collected between 56 and 217 days post symptoms onset by the German Red Cross - Blood Donation Service, Institute for Transfusion Medicine Dresden with the informed consent of blood donors and after consent vote of the institutional review board (EK138042014). 17 SARS-CoV-2 infected patients (5 female, 13 male) represented non-severe COVID-19 cases according to the classification of WHO. One COVID-19 patient was classified as severe COVID-19 case and hospitalized but did not require intensive care unit (ICU) care. Blood samples from 11 HI (6 female, 5 male) were collected before 2019. Blood samples form HI tested negative for SARS-CoV-2 S IgGs (2 female, 2 male) were collected before 2020.
2.2. Isolation and cryopreservation of PBMCs
Peripheral blood mononuclear cells (PBMCs) were isolated by density-gradient centrifugation using Ficoll-Hypaque as described. Briefly, heparinized whole blood were diluted with PBS/HSA (5%) (v/v) and carefully layered on Biocoll (Biochrom, Berlin, Germany) and centrifuged with 800 g for 30 min at RT. The white layer were isolated and washed with PBS/HSA by centrifugation 300 g and 5 min at 4 °C. PBMCs were frozen in X-Vivo10 (Lonza, Basel, Switzerland) with 10% DMSO (Sigma-Aldrich, St. Louis, US-MO) and 40 mg/ml HSA (Baxter, Unterschleißheim, Germany) using controlled rate freezing containers ‘Mr. Frosty’ (Nalgene Nunc Int., Rochester, US-NY) and stored in the gas phase of a liquid nitrogen tank. Cell counts and viability were obtained using Trypan blue staining (Thermo Fisher Scientific, Waltham, US-MA) and a TC20™ automated cell counter (Biorad).
2.3. Stimulation of PBMCs with SARS-CoV-2 and CMV pp65 peptide mixes
To detect virus-peptide-reactive CD3+ T cells approximately 1.5 × 106 PBMCs were stimulated with SARS-Cov-2 M, N, S and S1 (Miltenyi Biotec, Bergisch Gladbach, Germany) or CMV pp65 peptide mixes (JPT Peptide Technologies, Berlin, Germany) consisting of 15-mers overlapping by 11 amino acids at a concentration of 1 µM/per single peptide in RPMI1640 with 10 mg/ml HSA in a 96-well round bottom plate (Greiner Bio-one, Kremsmuenster, Austria) at 37 °C in a humidified atmosphere. In addition, αCD28 (clone: L293) (BD Biosciences) at a final concentration of 1.3 µg/ml and a total volume of 150 µl/well was added as a co-stimulatory signal to each well. As negative control PBMCs were stimulated with αCD28 alone. To enable detection of intracellular cytokines 1 h after stimulation with peptide mixes and aCD28 mAb, GolgiPlug™ (BD Biosciences) was added to every well followed by an additional 4 hs of incubation. Cells were then washed with PBS/HSA (5 mg/ml) and stained with Ethidium monoazide (EMA) (Thermo Fisher Scientific) prior to fixation with Cytofix/Cytoperm solution (BD Biosciences). Subsequently, monoclonal antibody staining was performed with αCD3 BV421 (clone: UCHT1), αCD4 APC-Vio770 (clone: M-T321) (Miltenyi Biotec), αCD8 V500 (clone: SK1), αIFN-γ FITC (clone: 25,723.11), αTNF-α PE-Cy7 (clone: Mab11) (all BD Biosciences) and αIL-2 PE (clone: MQ1–17H12) (eBioscience, San Diego, US). Gates were set according to FMO controls. T-cell assays, e.g. ICS, presented in this work were performed compliant to MIATA guidelines.
2.4. Flow cytometric analysis
For flow cytometric analysis were performed using a FACS Canto II equipped with three lasers (blue 488 nm, red 633 nm and violet 405 nm) and Diva-Software V6.1.3 (both BD Bioscience) and adhered to the guidelines for the use of flow cytometry and cell sorting in immunological studies [28]. PMT voltages were adjusted to yield optimal signal to noise ratios. Compensation was applied for each fluorochrome. Gating strategies and a representative data set are shown in Supplementary Fig. 1. Data analyses was performed using FlowJo software V9.3.2 (FlowJo LLC, Ashland, US-OR).
2.5. Quantification and statistical analysis
Statistical data were calculated using GraphPad Prism software V6.02 (GraphPad Software Inc., La Jolla, US-CA). For the identification and determination of the frequency of intracellularly cytokine producing, antigen-reactive T cells the gating strategy as displayed in Supplementary Figure 1 was applied. In order to evaluate the presence of SARS-CoV-2-specific T-cell immunity a Stimulation Index (SI) was calculated as following: percentage of intracellular cytokine positive T cells following stimulation with SARS-CoV-2 peptide mixes (n = 1) divided by the average (arithmetric) mean value of the percentage of intracellular cytokine positive T cells (from 3 replicative measurements, n = 3) following incubation with αCD28 alone plus its SD. T-cell responses with a SI were rated positive. The frequency of SARS-CoV-2- and CMV pp65-specific T cells immunity given in Figures and in Tables was determined by subtracting the average arithmetic mean frequency of intracellular cytokine positive T cells detected following three independent stimulation with aCD28 alone (n = 3) from the frequency of intracellular cytokine positive T cells detected following stimulation with peptide mixes. Statistical significance (p) of antigen-reactive T cells frequencies between various groups was calculated using Student´s unpaired t-test.
2.6. Virus neutralization assay
Plaque reduction neutralization test (PRNT) were performed as described before [29]. Briefly, 4 × 105 cells/ml VeroE6 cells were seeded in 24-well plates 1 day prior testing. Patient plasma was heat-inactivated at 56 °C for 30 min, diluted with OptiPro medium (Fisher scientific, Schwerte, Germany) starting from a titer of 1:20 to 1:640 and incubated with a solution containing 100 plaque forming units of SARS-CoV-2 at 37 °C for 1 h. The virus containing plasma were then added in duplicates to the wells of 24-well plates and incubated at 37 °C. After 1 h the supernatant was discarded and the cells were washed once with PBS and supplemented with 1.2% Avicel solution in DMEM (Merck, Darmstadt, Germany). After 3 days incubation at 37 °C the supernatants were removed and the cells were fixed with PBS containing 6% formaldehyde and stained with crystal violet as described [30].
2.7. Determination of SARS-CoV-2-S protein-specific IgG
SARS CoV-2 IgG titre was determined using a CE-marked anti-SARS-CoV-2 IgG semiquantitative ELISA (Euroimmun, Lübeck, Germany) according to the manufacturer´s instructions. Results are given as the ratio of the optical density of the patient sample divided by arbitrary unit ratio of the provided control sample antibody. Antibody ratios (AbR) of ≥0.8 were considered positive.
2.8. Detection of SARS-CoV-2-, SARS-CoV-1-, MERS-CoV- and HCoV-specific IgG using multiplex antibody bead array
In order to extend the characterization of the humoral response to additional SARS-CoV-2 antigens including the RBD, nucleocapsid (N), S1/S2 domain and to the S1 domain of various relevant HCoVs (HCoV-OC43, HCoV-HKU1, HCoV-229E and HCoV-NL63) the serum of reconvalescent COVID-19 patients was further assessed using the SARS-CoV-2 specific multiplex antibody detection array (MABA) (Labscreen COVID plus) (One lambda, West Hill, CA, USA) and HLA Fusion software (Luminex, Austin, TX, USA)). Briefly, prior detection of SARS-CoV-2-specific IgGs 1 µl EDTA (0.2 M) was added to 20 µl of diluted (1:10) human serum. 20 µl diluted human serum was incubated with 5 µl Labscreen COVID plus multiplex beads for 30 min at RT in the dark. Following washing three times with PBS/HSA buffer multiplex beads were resuspended with 100 µl human IgG-PE and incubated for additional 30 min at RT in the dark. After washing two times with PBS/HSA the multiplex beads were resuspended in 80 µl PBS and analysed using Luminex xMAPbased assay and a LABScan3D (Luminex, Austin, TX, USA) and Microsoft Excel. Cutoff values for each recombinant protein were calculated and given as stated by the manufacturer using the MFI + 3xSD of 96 COVID-19 negative samples collected before 2019 [31].
3. Results
3.1. SARS-CoV-2 M, N and S/S1 protein-reactive T-cell immunity
SARS-CoV-2-infections have been shown to trigger Th1 responses with specificity to the structural immunodominant viral M, N and S protein. The S1 subdomain comprises the RBD within the S protein. Therefore, the SARS-CoV-2-specific T-cell immunity was examined for the presence of T cells with reactivity to the M, N, S and S1 protein in convalescent COVID-19 patients by ICS for IL-2, IFN-γ and TNF-α following stimulation with SARS-CoV-2-peptide mixes.
In total, PBMCs of 18 convalescent COVID-19 patients were analysed for SARS-CoV-2-reactive T-cell immunity. To identify patients with SARS-CoV-2-reactive T-cell responses SI were calculated and patient with a SI 3 were rated to manifest SARS-CoV-2-reactive T-cell immunity (Supplementary Fig. 2). In 17 COVID-19 patients (94%) SARS-CoV-2- reactive IL-2 and/or IFN-γ producing CD4+ or CD8+ T cells were detected. CD4+ SARS-CoV-2- reactive immunity was found in 16 (89%) COVID-19 patients which was directed to the SARS-CoV-2 M (n = 11) (61%), N (n = 10) (56%), S (n = 13) (72%) or S1 protein (n = 12) (67%). In contrast, only 10 (56%) of these COVID-19 patients demonstrated SARS-CoV-2- reactive IL-2 or IFN-γ producing CD8+ T cells with a specificity for the SARS-CoV-2 M (n = 7) (39%), N (n = 7) (39%), S (n = 7) (39%) and S1 protein (n = 4) (22%).
Also in the majority of the tested HI SARS-CoV-2-reactive T-cell immunity was detectable. In 12 out of 15 HI (80%) SARS-CoV-2-reactive CD4+ T-cell immunity and in 9 (60%) CD8+ T-cell immunity reactive for at least one of the tested SARS-CoV-2 peptide mix was detected. Similar to the COVID-19 patients CD4+ T cells reactive for all tested SARS-CoV-2 peptide mixes without prevalence could be detected (M (n = 6) (40%), N (n = 5) (33%), S (n = 7) (47%) or S1 protein (n = 3) (20%). In contrast, CD8+ T-cell immunity showed predominant reactivity for SARS-CoV-2 N and S/S1 proteins M (n = 3) (20%), N (n = 5) (33%), S (n = 5) (33%) or S1 protein (n = 5) (33%).
Remarkably, the determination of the SI of SARS-CoV-2-reactive CD4+ and CD8+ T cells was hampered by an abnormal high background of spontaneously TNF-α producing T cells in several COVID-19 patients. This becomes evident in discrepant SI found for CMV pp65- reactive T cell population producing various cytokines (Supplementary Fig. 2). While in COVID-19 patients displaying a high frequency of spontaneously producing TNF-α a negative SI for TNFα+ T cells reactive for CMV pp65 was calculated, positive SI (3) and significant frequencies of IFN-γ and IL-2 producing T cells following stimulation with CMV pp65 peptide mix were determined. In general, intracellular TNF-α expression in T cells was associated with IFN-γ co-expression.
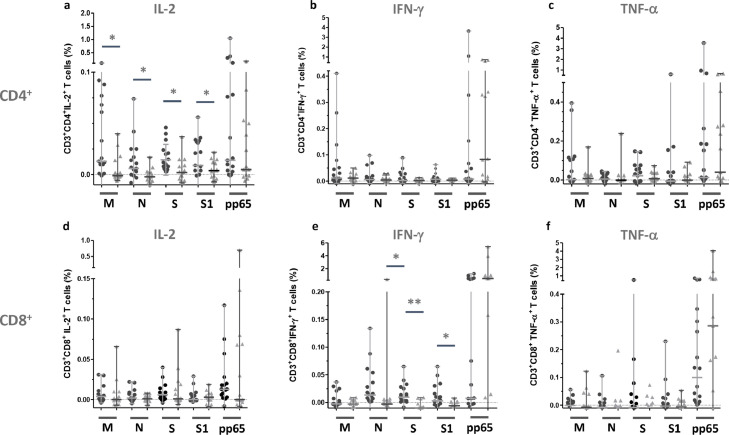
With regard to frequency the SARS-CoV-2-reactive T-cell immunity in the analysed COVID-19 convalescent patients is clearly dominated by CD4+ T cells reactive for the M protein. Median frequencies of 0.012% (ranging: −0.008%–0.410%) and 0.002% (ranging: −0.108%–0.395%) SARS-CoV-2 M protein-reactive IFN-γ+and TNF-α+CD4+ T cells in convalescent COVID-19 patients were observed (Fig. 1 b and c, Table 1 ). However, compared to the median frequencies of SARS-CoV-2 M protein-reactive IFN-γ+CD4+ T cells and TNF-α+CD4+ T cells detected in PBMCs from HI the median frequencies were not increased (Fig. 1b, Table 2 ). Also, median frequencies of N, S and S1 protein-reactive of IFN-γ+and TNF-α+CD4+ T cells detected in COVID-19 patients were not increased compared to the frequencies detected in HI (Fig. 1b and c, Table 1 and 2).
Fig. 1.
Frequency of SARS-CoV-2 M, N, S, S1 protein-reactive T cells detected in PBMCs of convalescent COVID-19 patients and healthy individuals.
Frequency of IFN-γ, IL-2 and TNF-α expressing CD4+or CD8+T cells detected after stimulation with respective SARS-CoV-2 M, N and S/S peptide mixes among T cells from convalescent COVID-19 patients and HI. Each closed black circles represent a single measurement of one convalescent COVID-19 patients. Each gray triangle represent a single measurement of one HI. Symbols and bars represent median and range. Statistically significant differences (p < 0.05) between groups of antigen-reactive T cells are indicated with horizontal bars and *. Highly statistical differences (p < 0.01) are indicated with horizontal bars and **.
Table 1.
Median frequency of SARS-CoV-2 protein-specific CD3+ T cells subpopulation in COVID-19 patients.
| Antigen | M | N | S | S1 | CMV pp65 | |
|---|---|---|---|---|---|---|
| CD4+ | IL-2 | 0.013 | 0.006 | 0.010 | 0.009 | 0.014 |
| IFN-γ | 0.012 | 0.006 | 0.002 | 0.007 | 0.008 | |
| TNF-α | 0.002 | 0.005 | 0.022 | −0.006 | 0.013 | |
| CD8+ | IL-2 | 0.004 | 0.002 | 0.006 | 0.000 | 0.013 |
| IFN-γ | −0.003 | 0.014 | 0.010 | 0.003 | 0.008 | |
| TNF-α | 0.004 | −0.007 | −0.008 | −0.003 | 0.133 |
Table 2.
Median frequency of SARS-CoV-2 Protein-specific CD3+ T cells subpopulation in HI (peripheral blood collected prior 2019).
| Antigen | M | N | S | S1 | CMV pp65 | |
|---|---|---|---|---|---|---|
| CD4+ | IL-2 | −0.001 | −0.002 | 0,002 | 0,004 | 0.005 |
| IFN-γ | 0.012 | 0.004 | 0.002 | 0.007 | 0.083 | |
| TNF-α | 0.008 | −0.001 | 0.007 | −0.001 | 0.040 | |
| CD8+ | IL-2 | −0.003 | −0.003 | 0.001 | 0.003 | 0.004 |
| IFN-γ | −0.003 | −0.003 | −0.01 | −0.006 | 0.482 | |
| TNF-α | −0.008 | −0.023 | −0.012 | −0.006 | 0.286 |
Statistically significant differences between frequencies of SARS-CoV-2 M, N and S protein- reactive IL-2+CD4+ T cells detected in COVID-19 patients and HI were found. The frequencies of SARS-CoV-2 M protein-reactive IL-2+CD4+ T cells detected in convalescent COVID-19 patients (median: 0.013%, range: −0.023%–0.108%) were significantly higher (p = 0.036) compared to the frequency found in HI (median: −0.001%, range: −0.006%–0.040%) (Fig. 1a, Tables 1 and 2). In addition, the frequency of SARS-CoV-2 N, S and S1 protein-reactive IL-2+CD4+ T cells detected in convalescent COVID-19 patients (median: 0.006%, range: −0.014%–0.074%) and (median: 0.010%, range: −0.014%–0.046%), (median: 0.009%, range: −0.023%–0.056%) (Table 1), respectively were significantly increased (p = 0.045 and p = 0.017, respectively) compared to HI demonstrating low frequencies of SARS-CoV-2 N, S and S1 protein-reactive IL-2+CD4+ T cells (median: −0.002%, range: −0.010%–0.017%), (median: 0.002%, range: −0.008%–0.037%) and (median: 0.004%, range: −0.007%–0.022%) (Fig. 1a, Table 2).
In comparison to the frequencies of SARS-CoV-2-reactive CD4+ T cells the detected frequencies of SARS-CoV-2-reactive CD8+ T cells are rather low (Figs. 1d-f). Moreover, in contrast to CD4+ T cells, SARS-CoV-2-reactive CD8+ T-cell immunity is dominated by IFN-γ+CD8+ T cells reactive for the N protein (median: 0.014%, range: −0.022%–0.134%) S and S1 protein (median: 0.010%, range: −0.075%−0.065%) (median: 0.003%, range: −0.054%–0.065%) (Fig. 1e, Table 1). Frequencies of IFN-γ+CD8+ T cells were statistically significant higher compared to those detected in HI (N: median: −0.003%, range: −0.078%–0.353%) (p = 0,011), (S: median: −0.010%, range: −0.070%−0.009%) (p = 0.003), and (S1 protein: median: −0.010%, range: −0.070%–0.009%) (p = 0.034) (Fig. 1e, Table 2), respectively.
Considerable relevant frequencies of SARS-CoV-2-reactive CD8+ T cells producing IL-2 were limited to convalescent COVID-19 patients (Fig. 1d). The median frequencies for SARS-CoV-2 M and S protein-reactive were 0.004% (range: −0.004–0.031) and 0.006% (range: −0.004–0.04) (Table 1).
CMV pp65-reactive T-cell immunity was detected with IL-2+ CD4+/IFN-γ+CD4+ and IL-2+ CD8+/IFN-γ+CD8+ T cells in eleven (61%) convalescent COVID-19 patients and in 11 out of 16 analysed (69%) HI (Fig. 1d and e). In five COVID-19 patients CMV pp65-reactive T cell immunity characterized by TNF-α production was disguised by high background of spontaneously TNF-α-producing T cells (Fig. 1f).
3.2. Significant increase of SARS-CoV-2 N and S protein-reactive T-cell immunity in convalescent patients up to 256 days after onset of disease
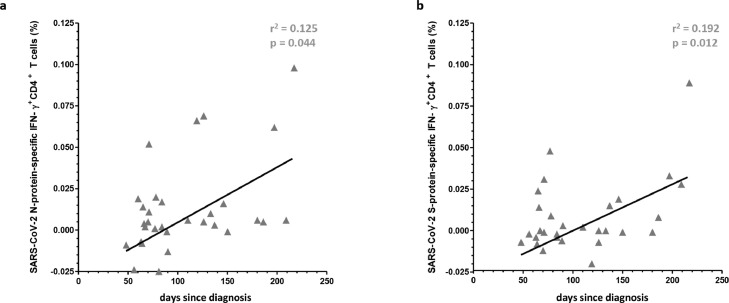
SARS-CoV-2-specific T cell immunity revealed a moderate increase and correlation of IFN-γ+CD4+ T-cells reactive for the SARSCoV-2 N- and S-protein (r2 = 0.125, p = 0.0044 and r2 = 0.192, p = 0.012) (Fig. 2 a and b) progressing with time following infection indicating generation and maintenance of a robust SARS-CoV-2-specific cellular immunity for these SARS-CoV-2 antigen in convalescent patients up to 256 days after diagnosis. No significant correlation was found between IFN-γ+CD4+ T cell immunity reactive for the SARSCoV-2 M- and S1-protein (data no shown).
Fig. 2.
Generation and maintenance of SARS-CoV-2-reactive IFN-γ+CD4+ T cells in con-valescent patients up to 256 days after onset of disease.
Frequency of SARS-CoV-2-N or S peptide mix-reactive IFN-γ+CD4+ T cells in convalescent patients up to 256 days after onset of disease, respectively (Fig. 2a and b). Regression lines, coefficient of variations (r2) and statistical significance (p-value) are depicted in the figures. Each symbol represents a single measurement.
3.3. Determination of humoral immunity towards SARS-CoV-2 and HCoVs
Anti-SARS-CoV-2-specific antibody responses were analysed with three different methods. Using the anti-SARS-CoV-2 S IgG ELISA relative amounts of serum IgG specific for the SARS-CoV-2 S protein, which comprises the RBD, in convalescent COVID-19 patients were determined. In addition, SARS-CoV-2 neutralizing antibodies were measured with the PRNT. MABA was used to detect the HCoV-OC43, HCoV-HKU1, HCoV-229E, HCoV-NL63 and SARS-CoV-2 S1 protein-specific IgGs including the RBD, N and S1/S2 protein of SARS-CoV-2. In order to evaluate the interrelation of the different assays used for the detection of humoral immune responses to SARS-CoV-2 and HCoVs the results from respective assays were analysed and compared. A significant correlation was found between the concentration of anti-SARS-CoV-2 S protein-specific IgGs determined by ELISA and anti-S1 protein-specific IgGs (r2 = 0.696, p = 0.0027) (Supplementary Fig. 3a) or anti-SARS-CoV-2 S (r2 = 0.739, p = 0.0014) (Supplementary Fig. 3b), detected by MABA with a more stringent correlation between IgGs specifically binding to the S protein.
Additionally, significant correlations were found between neutralizing SARS-CoV-2 antibodies and SARS-CoV-2 S protein-specific IgGs, detected by ELISA (r2 = 0.456, p = 0.0081) (Supplementary Fig. 4a) and anti-SARS-CoV-S1 protein-specific IgGs, detected by MABA (r2 = 0.290, p = 0.0026) (Supplementary Fig. 4b).
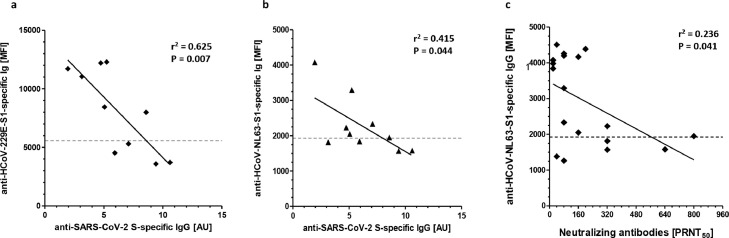
Interestingly, an inverse correlation was found between anti-HCoV-NL63 S1 and anti-HCoV-229E S1 protein-specific IgGs, detected by MABA, and anti-SARS-CoV S protein-specific IgGs, detected by ELISA (r2 = 0.415, p = 0.044 or r2 = 0.625, p = 0.007), respectively (Fig. 3 a and b). Furthermore, an inverse correlation was detected between anti-HCoV-NL63 S1 protein-specific IgGs and neutralizing antibodies (r2 = 0.236, p = 0.041) (Fig. 3c).
Fig. 3.
Correlation of anti-HCoV-NL63 S1/anti-HCoV-229E S1 protein-specific IgGs levels with anti-SARS-CoV-2 S/S1-specific IgGs levels and SARS-CoV-2 neutralizing antibody titer.
MFI values of anti-SARS-CoV-2 S1-specific IgGs levels determined by MABA were compared with MFI values of anti-HCoV-NL63 S1-protein-specific IgGs (Fig. 2a) or anti-HCoV-2-229E S1-protein-specific IgGs (Fig. 2b) also determined by MABA in sera of convalescent COVID-19 patients. The titer of SARS-CoV-2 neutralizing antibodies is correlated to MFI values of anti-HCoV-NL63 S1-protein-specific IgGs (Fig. 2c). Regression lines, coefficient of variations (r2) and statistical significance (p-value) are depicted in the Figures. Each symbol represents a single measurement.
3.4. Correlation of cellular and humoral immunity towards SARS-CoV-2 and HCoVs
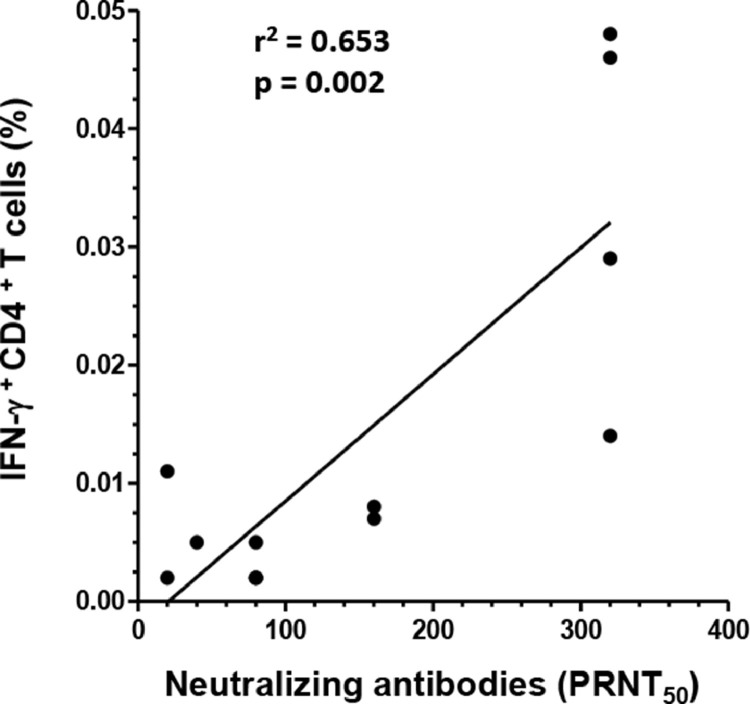
A strong correlation was found between the SARS CoV-2 S neutralizing antibody titers and the frequency of S1 protein-reactive IFN-γ+CD4+ T cells (r2 = 0.653, p = 0.002) (Fig. 4 ), respectively. In contrast, no relevant correlations were revealed between SARS-CoV-2 S protein- reactive IgG and M, N or S1protein-reactive IFN-γ+CD4+ T cells or S protein-reactive IL-2+CD4+ T cells (data not shown).
Fig. 4.
Correlation of SARS-CoV-2 neutralizing antibody titer with the frequency of anti-SARS-CoV-2 S1 protein-reactive INF-γ+CD4+ T cells detected in convalescent COVID-19 patients.
Titer of SARS-CoV-2 neutralizing antibodies were compared with the frequency (Fig. 3) of anti-SARS-CoV-2 S1 protein-reactive INF-γ+ CD4+ T cells detected in convalescent COVID-19 patients. Regression lines, coefficient of variations (r2) and statistical significance (p-value) are depicted in the figure. Each symbol represents a single measurement.
4. Discussion
In our study we investigated the nature of SARS-CoV-2-specific immunity in patients with mild COVID-19 and sought to identify parameters most relevant for the generation and maintenance of neutralizing antibody responses in convalescent COVID-19 patients. The cohort of convalescent COVID-19 patients analysed and presented here comprises 18 relative young individuals with PCR-confirmed SARS-CoV-2 infection. These COVID-19 patients except one showed mild disease symptoms without the requirement of hospitalization. After recovery from SARS-CoV-2 infection they were selected as potential donors for convalescent plasma containing anti-SARS-CoV2 neutralizing antibodies according to the amount of anti-SARS-CoV-2-specific IgGs detected in their serum.
In order to investigate the correlation of the humoral response against SARS-CoV-2 and various `common cold´ HCoVs we extended our analysis using MABA. The SARS-CoV-2-reactive T-cell immunity in the examined cohort of convalescent patients was investigated using ICS following provocation with SARS-CoV-2 M, N, S and S1 protein-specific peptide mixes and compared to HI who donated blood before the beginning of the pandemic.
Measurements of the SARS-CoV-2-specific humoral immunity using various assays gave comparable results with statistically significant correlations. Highly significant correlation between the levels of SARS-CoV-2 S-protein-specific IgGs detected by ELISA and MABA showed the good comparability of both methods. Slightly reduced correlation coefficient of SARS-CoV-2 S protein-specific IgG and SARS-CoV-2 S1 protein-specific confirmed presence of SARS-CoV-2 S1 protein-specific IgGs in convalescent COVID-19 patients. However, a significant amount of SARS-CoV-2 S protein-specific IgGs are directed to the S1 protein. Importantly, the neutralizing capacity of detected SARS-CoV-2 S/S1 protein-specific IgG was demonstrated by significant correlation of the results yielded by the PRNT. The comparison of the SARS-CoV-2 S protein-specific IgG values also indicates a certain degree of variation in the detection of SARS-CoV-2-specific epitopes, particularly with respect to the RBD.
While strong antibody responses to MERS and SARS-CoV-2 coincide with the clinical course of the disease, magnitude and character of the T-cell immunity had been reported being less affected by the severity of the disease and SARS-CoV-2-specific T-cell immunity present in the majority of COVID-19 patients ([7, 18, 32, 33]). Since neutralizing, durable antibody responses and affinity-matured B cell memory depend on CD4+ T-cell help [34], key to the understanding of humoral immune response directed towards SARS-CoV-2 is the elucidation of T-cell immunity and epitopes recognized by CD4+ T cells.
Several publications described SARS-CoV-2 epitopes recognized by CD4+ and CD8+ T cells after infection and reported that the majority of CD4+ T-cell mediated SARS-CoV-2-specific immunity is directed towards structural proteins with the highest frequencies towards S/S1, M and N proteins ([33, 35, 36]). In our study we detected comparatively high frequencies M and N protein-specific and low frequencies of S/S1 protein-reactive CD4+ T cells possibly due to the examined patient cohort exhibiting mild COVID-19. However, following up the SARS-CoV-2 reactive T-cell immunity we were able to detect an increase and statistically significant correlation of IFN-γ+CD4+ T-cell immunity reactive for SARS-CoV-2 N- and S-protein in the analysed COVID-19 patients over time indicating generation and maintenance of SARS-CoV-2-specific cellular immunity in convalescent patients for more than 8 months (256 days) after onset of disease. This has been previously shown by Mazzoni et al., ([37, 38]). Of note, CD4+ T-cell immunity following infection with common cold HCoVs had been reported being predominantly directed towards the S protein [17], whereas convalescent MERS patients show a more diverse T-cell immunity directed towards M, N, and S protein [39].
In comparison to CD4+ T cells, SARS-CoV-2-specific CD8+ T-cell immunity has been described to be preferentially directed to S and N protein following infections with SARS-CoV or SARS-CoV-2. A striking predominance of S protein-reactive CD8+ T-cell immunity was found in patients infected with SARS-CoV. There, S protein-reactive CD8+ T cells amounted to approximately 50% of total detected SARS-CoV-reactive T cells whereas N protein-specific CD8+ T cells accounted for 36% [17] SARS-CoV-reactive CD8+ T cells. A similar finding was reported by Thieme et al. analysing T-cell immunity in COVID-19 patients [33]. A predominant S and N protein-reactive CD8+ T-cell immunity following SARS-CoV-2 infection was described for COVID-19, which is in good agreement with our findings presented here. However, studies analysing the T-cell immunity in COVID-19 patients reported that CD8+ T-cell immunity were predominantly directed to the N protein or M, N, and S protein without precedence ([33, 40]).
With respect to the magnitude of SARS-CoV-2-reactive T-cell immunity we detected markedly higher CD4+ T-cell responses in terms of frequency compared to CD8+ mediated T-cell responses. Also, this finding is in good agreement with various other studies ([33, 35, 41]).
Furthermore, we could confirm the presence of SARS-CoV-2-reactive T-cell immunity in the vast majority of tested convalescent COVID-19 patients. This is in line with the study of Grifoni et al. reporting on the presence of SARS-CoV-2-reactive CD4+ T cells in 100% and CD8+ T cells in 70% of tested COVID-19 patients [35]. Other studies stated lower proportions for various reasons [33]. However, the majority of the studies demonstrated that the predominant portion of tested COVID-19 patients revealed robust SARS-CoV-2-reactive T-cell immunity [37], even in absence of SARS-CoV-2-specific antibody [42]. Observed discrepancies between these findings can be possibly attributed to relative small sample numbers, different T-cell stimulation and staining methods and variable cell activation marker (CD137, CD154, CD69 or intracellular cytokines) used for the identification of SARS-CoV-2-reactive T cells.
While SARS-CoV and MERS-reactive T-cell immunity was confirmed to be persistent for many years following infection humoral immunity waned within months and virus-specific antibodies are not detectable in the majority of the virus-infected individuals after 2–3 years. For the induction of high affinity antibody responses and persistent humoral immunity CD4+ T-cell help is required. Therefore, evidence for correlation of SARS-CoV-2-reactive CD4+ T-cell immunity and SARS-CoV-2-specific humoral immunity can be taken as a strong indicator for the induction of a robust and durable SARS-CoV-2-specific humoral immunity. Our finding, that there is a significant correlation between SARS-CoV-2 neutralizing antibody titers and S/S1 protein-reactive IFN-γ+CD4+ T cells, provides further evidence for the induction of a robust humoral immunity directed to the S/S1 protein of SARS-CoV-2 in convalescent COVID-19 patients as described before ([37, 38, 40]).
Remarkably, the presence anti-HCoV-NL63 and anti-HCoV-229E S1 protein-specific IgGs is inversely correlated with the induction of humoral immunity directed to the SARS-CoV-2 S protein suggesting a potential suppressive impact of preexisting anti-HCoV-NL63 and anti-HCoV-229E S1 protein-specific IgG on the induction of humoral immunity specific for SARS-CoV-2. A negative impact of pre-existing humoral immunity specific for common cold HCoVs on the induction of SARS-CoV-2 specific antibody responses has been recently published. Aydillo et al., reported that antibodies specific for the conserved region of the common cold HCoVs spike protein were boosted in COVID-19 patients which in turn had a negative impact on the induction of antibodies against SARS-CoV-2 S and N proteins [43].
However, no such immunological imprint on the induction of SARS-CoV-2-specific antibodies was observed for HCoV-229E. HCoV-NL63 was not analysed in this analysis. In another report Lin et al. described that pre-existing humoral immunity to HCoVs S protein SARS-CoV-2 impedes the generation of SARS-CoV-2 neutralizing antibodies in mice [44].
The inhibitory effect of existing anti-HCoV-NL63 and anti-HCoV-229E S1 protein-specific IgGs may result from the suppression of activation of naive B cells and their subsequent proliferation and differentiation to plasma cells when a simultaneous binding of the antigen in the immune complex to the B cell receptor and to the inhibitory Fc-γRIIB occurs [45]. A well-known example is the failure of seroconversion of babies when vaccinated against measles within 6 months after birth as a result of the acquisition of MV-specific antibodies via transplacental transfer of maternal IgGs. Since maternal IgGs have limited half-live MV vaccinations later are much more efficient [46]. Likewise, investigations in cynomolgus macaques showed that even low pre-existing antibody titers can inhibit the generation of antibodies following vaccination with MV or recombinant vaccinia virus vector expressing measles antigens [47]. Furthermore, cross-linking of Fc-γRIIB induces apoptosis in bone marrow plasma cells [48] which consequently results in decreasing antibody levels. In addition, engagement of Fc-γRIIB by IgG immune complexes led to the failure of DC maturation resulting in inefficient CD4+ T-cells support required for B-cell activation and plasma cell differentiation [49].
Of note, the levels of S1 protein-specific IgGs of HCoV-OC43 and HCoV-HKU1 sharing a higher homology with SARS-CoV-2 [50] than HCoV-229E, were found to be clearly lower compared to S1 protein-specific IgG levels of HCoV-229E. The higher concentration of protein-specific IgG levels of HCoV-229E might have contributed to the contradicting finding concerning the inhibitory effect of pre-existing antibodies specific for the S protein of common cold Alpha- and Betacoronavirus.
Whether cross-reactive HCoVs-specific immunity is beneficial or detrimental with regard to the susceptibility to SARS-CoV-2 infection or severity of COVID-19 still remains elusive. Recent findings indicate that pre-existing immunity to seasonal coronaviruses may increase the susceptibility to SARS-CoV-2 [51]. In the context of convalescent plasma transfusion with its proven clinical benefit for critically ill COVID-19 patients ([52, 53]) the presence of anti-HCoV-NL63 and anti-HCoV-229E IgG might have an adverse impact on the therapy outcome and should therefore be considered.
5. Limitation of study
Using peptide mixes covering M, N, S and its subdomain S1 protein bears the risk that a large portion of potential SARS-CoV-2-reactive T cells are missed by the analysis. Low frequency of detected SARS-CoV-2-reactive T cells accompanied by an unusual high background staining – particularly for TNF-α expressing CD8+ T cells – hampered the analysis of SARS-CoV-2-reactive cellular immunity in many cases. Therefore, we confined our study to the analysis of IL-2 and IFN-γ. Although previous studies confirmed the predominant Th1 nature of the SARS-CoV-2 T-cell immunity ([36, 41, 54]), particularly directed to SARS-CoV-2 S protein in patients with mildly symptomatic COVID-19 after three months of infection [55] - the detected T-cell reactivity may not represents all facets of SARS-CoV-2-specific T-cell immunity in convalescent COVID-19 patients.
Testing of the humoral immune response specific for SARS-CoV-2 M protein was not performed due to the limitations of available test kits. Considering the predominance of SARS-CoV-2 M protein- reactive CD4+ T cells found in COVID-19 patients our study might not fully assess the role of M protein-specific humoral immunity in the context of SARS-CoV-2 infection.
In addition, the small cohort of convalescent patients of relative young age exhibiting mild to moderate COVID-19 may have contributed to biasing the detected humoral and cellular SARS-CoV-2-specific immunity. Further studies should address the issue how pre-existing, cross-reactive immunity from prior infections with common cold HCoVs have an impact on SARS-CoV-2 infection and immunity.
6. Conclusion
In the majority of the examined mild COVID-19 patients and HIs recruited before 2019 an immune response directed to various SARS-CoV-2 structural proteins was detected. The finding of an anti-SARS-CoV-2-reactive cellular immune response in HI suggests a pre-existing immunity to common cold HCoVs which share homology with SARS-CoV-2. The humoral immunity to the S protein of SARS-CoV-2 detected in convalescent COVID-19 patients positively correlates with the presence of SARS-CoV-2-reactive CD4+ T cells expressing Th1 cytokines indicating the generation of a robust cellular and humoral SARS-CoV-2-directed immunity. The inverse correlation of SARS-CoV-2 S protein-specific IgGs with HCoV-NL63 and HCoV-229E S1 protein-specific IgGs suggest that pre-existing humoral immunity to common cold HCoVs, particularly Alphacoronaviruses, may have had an inhibitory imprint on the humoral immune response to SARS-CoV-2 infection detected in patients with mild COVID-19.
Supplementary Information
Supplementary Figure 1. Gating strategy for the identification of SARS-CoV-2 and CMV pp65 peptide-reactive T cells.
Intracellular IFN-γ, IL-2 and TNF-α expressing T cells following stimulation with SARS-CoV-2 M, N, S/S1 and CMV pp65 peptide-mixes were identified by the use of sequential and Boolean gates. Single viable lymphocytes were identified by their characteristic light scatter properties and relative lack of EMA staining (Supplementary Fig. 1a-c). CD4+ T cells and CD8+ T cells were identified by applying a Boolean gate according to the differential expression of CD4 and CD8 on CD3+ T cells (Supplementary Fig. 1d and e). The frequency of IFN-γ, IL-2 and TNF-α expressing CD4+ (Supplementary Fig. 1 f-h) and CD8+ T cells (Supplementary Fig. 1 i-k) were determined as depicted. Lower gate limits were set according to the staining of unstimulated T cells. Data from a representative flow cytometric analysis of PBMCs from a convalescent COVID-19 patient following stimulation with SARS-CoV-2 M peptide mix is shown.
Supplementary Figure 2: SI of SARS-CoV-2 M, N, S/S1 protein-reactive T cells detected in PBMCs of convalescent COVID-19 patients and HIs.
SI of IFN-γ, IL-2 and TNF-α expressing CD4+ or CD8+T cells detected after stimulation with respective SARS-CoV-2 M, N and S/S1 protein peptide mixes among CD3+ T cells of PBMCs from convalescent COVID-19 patients and HIs. Each symbol depicted represents one measurement. Symbols and bars represent median and range.
Supplementary Figure 3: Correlation of anti-SARS-CoV-2 S/S1 protein-specific IgGs levels determined using ELISA and MABA.
Anti-SARS-CoV-2 S1 protein-specific IgGs from the serum of convalescent COVID-19 patients were determined using MABA and MFI values were plotted against anti-SARS-CoV-2 S protein-specific antibody ratios (AbR) of IgGs detected by ELISA (Supplementary Fig. 3a). MFI values of anti-SARS-CoV-2 S protein-specific IgGs of convalescent COVID-19 patients were analysed using MABA and plotted against anti-SARS-CoV-2 S protein-specific IgGs antibody ratios (AbR) detected by ELISA. Regression lines, coefficient of variations (r2) and statistical significance (p-value) are shown. Each symbol represents a single measurement.
Supplementary Figure 4: Correlation of anti-SARS-CoV-2 S/S1 protein-specific IgGs levels using MABA and SARS-CoV-2 neutralizing antibody titers.
SARS-CoV-2 neutralizing antibodies were determined by PRNT and compared to anti-SARS-CoV-2 S-protein-specific IgGs antibody ratios (AbR) (Supplementary Fig. 4a) and to the MFI values of anti-SARS-CoV-2 S1 protein-specific IgGs (Supplementary Fig. 4b) determined using MABA in convalescent COVID-19 patients. Regression lines, coefficient of variations (r2) and statistical significance (p-value) are given in the figures. Each symbol represents a single measurement.
Table 1 Frequency of SARS-CoV-2 protein-reactive CD4+ and CD8+ T cells in convalescent COVID-19 patients.
Median frequencies of SARS-CoV-2 M, N, S/S1 protein and CMV pp65-reactive IFN-γ+/IL-2+/TNF-α+CD4+ T cells and CD8+ T cells detected in PBMCs of HI.
Table 2 Frequency of SARS-CoV-2 protein-reactive CD4+ and CD8+ T cells in HIs.
Median frequencies of SARS-CoV-2 M, N, S/S1 protein and CMV pp65-reactive IFN-γ+/IL-2+/TNF-α+CD4+ T cells and CD8+ T cells detected in PBMCs of HI.
Declaration of Competing Interest
All authors declare no conflict of interest.
Acknowledgements
We would like to acknowledge Matthias Johnsen, Martina Wohsmann, Knut Gubbe, Undine Schulz, Madeleine Teichert, Kerstin Frank and Victor M. Corman for excellent technical support.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.imlet.2022.09.007.
Appendix. Supplementary materials
References
- 1.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94(7) doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perlman S., Dandekar A.A. Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 2005;5(12):917–927. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng O.W., Chia A., Tan A.T., Jadi R.S., Leong H.N., Bertoletti A., et al. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine. 2016;34(17):2008–2014. doi: 10.1016/j.vaccine.2016.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020 doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 6.Cao W.C., Liu W., Zhang P.H., Zhang F., Richardus J.H. Disappearance of antibodies to SARS-associated coronavirus after recovery. N. Engl. J. Med. 2007;357(11):1162–1163. doi: 10.1056/NEJMc070348. [DOI] [PubMed] [Google Scholar]
- 7.Liu W., Fontanet A., Zhang P.H., Zhan L., Xin Z.T., Baril L., et al. Two-year prospective study of the humoral immune response of patients with severe acute respiratory syndrome. J. Infect. Dis. 2006;193(6):792–795. doi: 10.1086/500469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Callow K.A., Parry H.F., Sergeant M., Tyrrell D.A. The time course of the immune response to experimental coronavirus infection of man. Epidemiol. Infect. 1990;105(2):435–446. doi: 10.1017/s0950268800048019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang F., Quan Y., Xin Z.T., Wrammert J., Ma M.J., Lv H., et al. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J. Immunol. 2011;186(12):7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- 10.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang F., Nie J., Wang H., Zhao Q., Xiong Y., Deng L., et al. Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. J. Infect. Dis. 2020;221(11):1762–1769. doi: 10.1093/infdis/jiaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng H.Y., Zhang M., Yang C.X., Zhang N., Wang X.C., Yang X.P., et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol. Immunol. 2020;17(5):541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thevarajan I., Nguyen T.H.O., Koutsakos M., Druce J., Caly L., van de Sandt C.E., et al. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat. Med. 2020;26(4):453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol. Immunol. 2020;17(5):533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li C.K., Wu H., Yan H., Ma S., Wang L., Zhang M., et al. T cell responses to whole SARS coronavirus in humans. J. Immunol. 2008;181(8):5490–5500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ni L., Ye F., Cheng M.L., Feng Y., Deng Y.Q., Zhao H., et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52(6):971–977. doi: 10.1016/j.immuni.2020.04.023. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trivedi S.U., Miao C., Sanchez J.E., Caidi H., Tamin A., Haynes L., et al. Development and evaluation of a multiplexed immunoassay for simultaneous detection of serum IgG antibodies to six human coronaviruses. Sci. Rep. 2019;9(1):1390. doi: 10.1038/s41598-018-37747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agnihothram S., Gopal R., Yount B.L., Jr., Donaldson E.F., Menachery V.D., Graham R.L., et al. Evaluation of serologic and antigenic relationships between middle eastern respiratory syndrome coronavirus and other coronaviruses to develop vaccine platforms for the rapid response to emerging coronaviruses. J. Infect. Dis. 2014;209(7):995–1006. doi: 10.1093/infdis/jit609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao X., Zhou H., Wu C., Xiao Y., Ren L., Paranhos-Baccala G., et al. Antibody against nucleocapsid protein predicts susceptibility to human coronavirus infection. J. Infect. 2015;71(5):599–602. doi: 10.1016/j.jinf.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macnaughton M.R., Madge M.H., Reed S.E. Two antigenic groups of human coronaviruses detected by using enzyme-linked immunosorbent assay. Infect. Immun. 1981;33(3):734–737. doi: 10.1128/iai.33.3.734-737.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan K.H., Cheng V.C., Woo P.C., Lau S.K., Poon L.L., Guan Y., et al. Serological responses in patients with severe acute respiratory syndrome coronavirus infection and cross-reactivity with human coronaviruses 229E, OC43, and NL63. Clin. Diagn. Lab. Immunol. 2005;12(11):1317–1321. doi: 10.1128/CDLI.12.11.1317-1321.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehmann C., Wolf H., Xu J., Zhao Q., Shao Y., Motz M., et al. A line immunoassay utilizing recombinant nucleocapsid proteins for detection of antibodies to human coronaviruses. Diagn. Microbiol. Infect. Dis. 2008;61(1):40–48. doi: 10.1016/j.diagmicrobio.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Che X.Y., Qiu L.W., Liao Z.Y., Wang Y.D., Wen K., Pan Y.X., et al. Antigenic cross-reactivity between severe acute respiratory syndrome-associated coronavirus and human coronaviruses 229E and OC43. J. Infect. Dis. 2005;191(12):2033–2037. doi: 10.1086/430355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patrick D.M., Petric M., Skowronski D.M., Guasparini R., Booth T.F., Krajden M., et al. An outbreak of human coronavirus OC43 infection and serological cross-reactivity with SARS coronavirus. Can. J. Infect. Dis. Med. Microbiol. 2006;17(6):330–336. doi: 10.1155/2006/152612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aburizaiza A.S., Mattes F.M., Azhar E.I., Hassan A.M., Memish Z.A., Muth D., et al. Investigation of anti-middle East respiratory syndrome antibodies in blood donors and slaughterhouse workers in Jeddah and Makkah, Saudi Arabia, fall 2012. J. Infect. Dis. 2014;209(2):243–246. doi: 10.1093/infdis/jit589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cossarizza A., Chang H.D., Radbruch A., Acs A., Adam D., Adam-Klages S., et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies (second edition) Eur. J. Immunol. 2019;49(10):1457–1973. doi: 10.1002/eji.201970107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 30.Herzog P., Drosten C., Muller M.A. Plaque assay for human coronavirus NL63 using human colon carcinoma cells. Virol. J. 2008;5:138. doi: 10.1186/1743-422X-5-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bray R.A., Lee J.H., Brescia P., Kumar D., Nong T., Shih R., et al. Development and validation of a multiplex, bead-based assay to detect antibodies directed against SARS-CoV-2 proteins. Transplantation. 2021;105(1):79–89. doi: 10.1097/TP.0000000000003524. [DOI] [PubMed] [Google Scholar]
- 32.Lynch K.L., Whitman J.D., Lacanienta N.P., Beckerdite E.W., Kastner S.A., Shy B.R., et al. Magnitude and kinetics of anti-severe acute respiratory syndrome coronavirus 2 antibody responses and their relationship to disease severity. Clin. Infect. Dis. 2021;72(2):301–308. doi: 10.1093/cid/ciaa979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thieme C.J., Anft M., Paniskaki K., Blazquez-Navarro A., Doevelaar A., Seibert F.S., et al. Robust T cell response toward spike, membrane, and nucleocapsid SARS-CoV-2 proteins is not associated with recovery in critical COVID-19 patients. Cell Rep. Med. 2020;1(6) doi: 10.1016/j.xcrm.2020.100092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Follicular Crotty S.T. Helper cell biology: a decade of discovery and diseases. Immunity. 2019;50(5):1132–1148. doi: 10.1016/j.immuni.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501. doi: 10.1016/j.cell.2020.05.015. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohen K.W., Linderman S.L., Moodie Z., Czartoski J., Lai L., Mantus G., et al. Longitudinal analysis shows durable and broad immune memory after SARS-CoV-2 infection with persisting antibody responses and memory B and T cells. Cell Rep. Med. 2021;2(7) doi: 10.1016/j.xcrm.2021.100354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazzoni A., Maggi L., Capone M., Spinicci M., Salvati L., Colao M.G., et al. Cell-mediated and humoral adaptive immune responses to SARS-CoV-2 are lower in asymptomatic than symptomatic COVID-19 patients. Eur. J. Immunol. 2020;50(12):2013–2024. doi: 10.1002/eji.202048915. [DOI] [PubMed] [Google Scholar]
- 38.Mazzoni A., Maggi L., Capone M., Vanni A., Spinicci M., Salvati L., et al. Heterogeneous magnitude of immunological memory to SARS-CoV-2 in recovered individuals. Clin. Transl. Immunol. 2021;10(5):e1281. doi: 10.1002/cti2.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao J., Alshukairi A.N., Baharoon S.A., Ahmed W.A., Bokhari A.A., Nehdi A.M., et al. Recovery from the Middle East respiratory syndrome is associated with antibody and T-cell responses. Sci. Immunol. 2017;2(14) doi: 10.1126/sciimmunol.aan5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21(11):1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5(48) doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sekine T., Perez-Potti A., Rivera-Ballesteros O., Stralin K., Gorin J.B., Olsson A., et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183(1):158–168. doi: 10.1016/j.cell.2020.08.017. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aydillo T., Rombauts A., Stadlbauer D., Aslam S., Abelenda-Alonso G., Escalera A., et al. Immunological imprinting of the antibody response in COVID-19 patients. Nat. Commun. 2021;12(1):3781. doi: 10.1038/s41467-021-23977-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin C.-.L., Wolf J., Brice D.C., Sun Y., Locke M., Cherry S., et al. Pre-existing humoral immunity to human common cold coronaviruses negatively impacts the protective SARS-CoV-2 antibody response. Cell Host Microbes. 2021 doi: 10.1016/j.chom.2021.12.005. preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tzeng S.J., Li W.Y., Wang H.Y. FcgammaRIIB mediates antigen-independent inhibition on human B lymphocytes through Btk and p38 MAPK. J. Biomed. Sci. 2015;22:87. doi: 10.1186/s12929-015-0200-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Albrecht P., Ennis F.A., Saltzman E.J., Krugman S. Persistence of maternal antibody in infants beyond 12 months: mechanism of measles vaccine failure. J. Pediatr. 1977;91(5):715–718. doi: 10.1016/s0022-3476(77)81021-4. [DOI] [PubMed] [Google Scholar]
- 47.van Binnendijk R.S., Poelen M.C., van Amerongen G., de Vries P., Osterhaus A.D. Protective immunity in macaques vaccinated with live attenuated, recombinant, and subunit measles vaccines in the presence of passively acquired antibodies. J. Infect. Dis. 1997;175(3):524–532. doi: 10.1093/infdis/175.3.524. [DOI] [PubMed] [Google Scholar]
- 48.Xiang Z., Cutler A.J., Brownlie R.J., Fairfax K., Lawlor K.E., Severinson E., et al. FcgammaRIIb controls bone marrow plasma cell persistence and apoptosis. Nat. Immunol. 2007;8(4):419–429. doi: 10.1038/ni1440. [DOI] [PubMed] [Google Scholar]
- 49.Bournazos S., DiLillo D.J., Ravetch J.V. The role of Fc-FcgammaR interactions in IgG-mediated microbial neutralization. J. Exp. Med. 2015;212(9):1361–1369. doi: 10.1084/jem.20151267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang A.T., Garcia-Carreras B., Hitchings M.D.T., Yang B., Katzelnick L.C., Rattigan S.M., et al. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat. Commun. 2020;11(1):4704. doi: 10.1038/s41467-020-18450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wratil P.R., Schmacke N.A., Karakoc B., Dulovic A., Junker D., Becker M., et al. Evidence for increased SARS-CoV-2 susceptibility and COVID-19 severity related to pre-existing immunity to seasonal coronaviruses. Cell Rep. 2021 doi: 10.1016/j.celrep.2021.110169. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tonn T., Corman V.M., Johnsen M., Richter A., Rodionov R.N., Drosten C., et al. Stability and neutralising capacity of SARS-CoV-2-specific antibodies in convalescent plasma. Lancet Microbe. 2020;1(2):e63. doi: 10.1016/S2666-5247(20)30037-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodionov R.N., Biener A., Spieth P., Achleitner M., Holig K., Aringer M., et al. Potential benefit of convalescent plasma transfusions in immunocompromised patients with COVID-19. Lancet Microbe. 2021;2(4):e138. doi: 10.1016/S2666-5247(21)00030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183(4):996–1012. doi: 10.1016/j.cell.2020.09.038. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodda L.B., Netland J., Shehata L., Pruner K.B., Morawski P.A., Thouvenel C.D., et al. Functional SARS-CoV-2-specific immune memory persists after mild COVID-19. Cell. 2021;184(1):169–183. doi: 10.1016/j.cell.2020.11.029. e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.