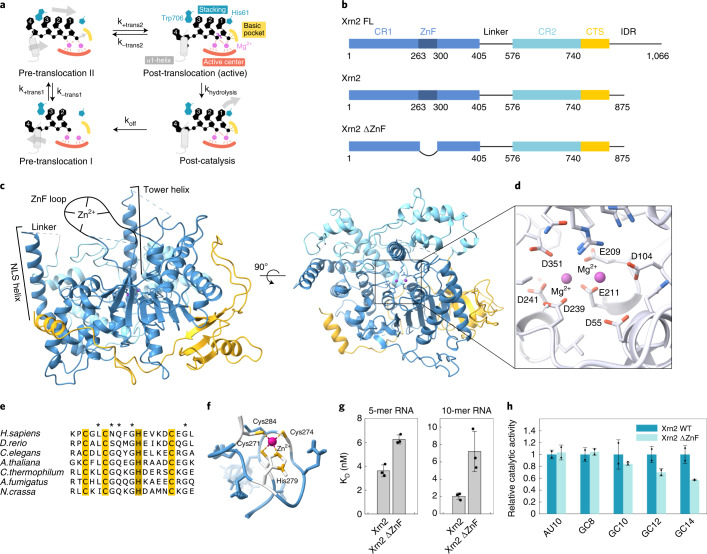
Fig. 1. Structure of the Xrn2 enzyme from the thermophilic eukaryote C. thermophilum.
a, The Xrn1/Xrn2 enzyme sandwiches the first three bases of the substrate RNA between a Trp and a His residue (top left; pre-translocation state II). Subsequently, the enzyme undergoes a conformational change and adopts the active conformation (top right). After hydrolysis (bottom right) and product release (bottom left), the substrate moves one base farther (top left). b, Schematic representation of the domain architecture of the full-length Xrn2 protein (top) and two constructs (Xrn2: middle; Xrn2 ΔZnF: bottom) used in this study. Solid boxes represent folded regions, whereas the linker region between CR1 and CR2 as well as the C-terminal intrinsically disordered region (IDR) are predicted to be unstructured and represented as lines. c, Crystal structure of CtXrn2, color-coded according to b. CR1 and CR2 and the Xrn2-specific CTS form a globular Xrn-core, in which the active site is accessible only from the top. d, Close-up of the active site, where seven conserved acidic residues (shown as sticks) coordinate two Mg2+ ions. e, Sequence alignment of the Xrn2 ZnF motif. The residues that coordinate the Zn2+ ion are highlighted in yellow (cysteines) and brown (histidine); other conserved residues are indicated with an asterisk. f, NMR structure of the ZnF region. g, Affinities of 5-mer and 10-mer RNA to WT Xrn2 and Xrn2 ΔZnF. Note that the binding experiments were performed in the absence of Mg2+ and in the presence of Zn2+. Data are shown as mean ± s.d. and were performed as triplicates. h, Relative degradation rates of Xrn2 WT and Xrn2 ΔZnF for different RNAs (Supplementary Table 4) containing either a 10-nucleotide AU hairpin (AU10) or GC hairpins with 8, 10, 12 or 14 nucleotides (GC8, GC10, GC12 and GC14). Data are shown as mean ± s.d. and were performed as two biological duplicates.