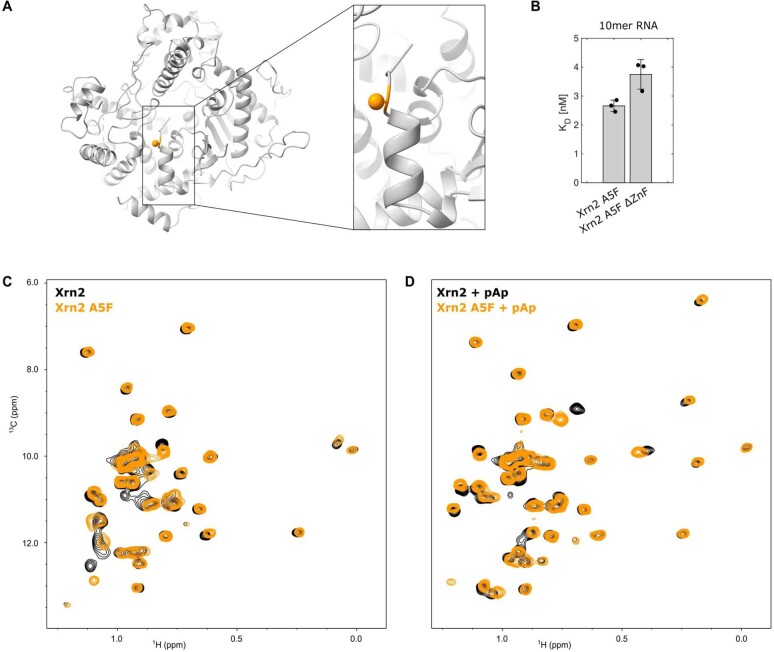
Extended Data Fig. 9. The A5F mutation does not interfere with the Xrn2 structure or with the binding of the substrate to Xrn2.
(A) Structure of Xrn2 with the mutation site A5 highlighted in orange and the Cβ atom depicted as a sphere. (B) Xrn2 A5F interacts with a 10mer RNA with very similar affinities as the WT enzyme (compare: Fig. 1G of the main text). Data points are shown as mean ± s.d., as derived from 3 independent experiments. (C) HMQC spectra of the Xrn2 WT (black) and Xrn2 A5F enzyme (orange). (D) HMQC spectra of the Xrn2 WT:pAp complex (black) and Xrn2 A5F:pAp complex (orange). Minor CSPs are visible around the N-terminal α1-helix, that are due to the A5F point mutation. The overall structural of the active state is, however, maintained in the presence of the A5F mutation (the black and orange spectra are highly similar in the absence and presence of substrate).