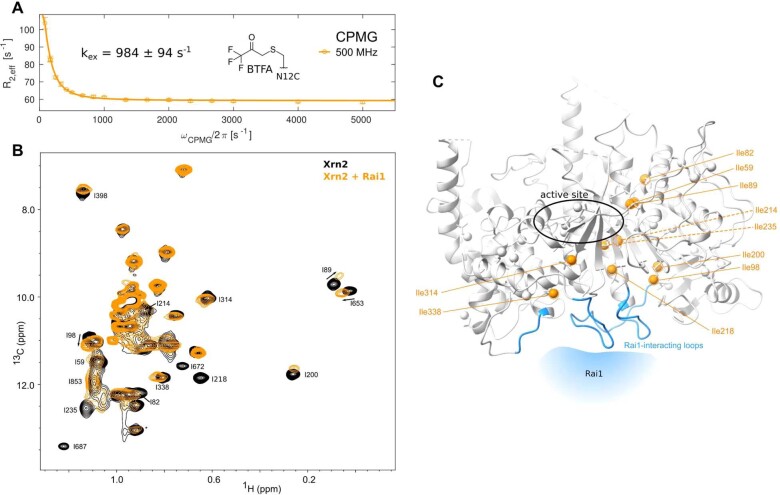
Extended Data Fig. 10. Changes in the Xrn2 enzyme spread from the Rai1 interface along the central β-sheet.
(A) 19F CPMG RD profile of Xrn2 ΔZnF N12CBTFA:Rai1 sample at 313 K. The fit yields an exchange rate of kex = 984 ± 94 s−1, which matches the exchange rate of Xrn2 ΔZnF N12CBTFA in the absence of Rai1 (kex = 913 ± 108 s−1). Binding of Rai1 thus does not influence the motions in Xrn2. Data points are shown as mean ± s.d., as derived from 3 duplicate NMR measurements. (B) HMQC spectrum of Ileδ1-[13CH3] and Metε-[13CH3] methyl labeled Xrn2 in the absence (black) and presence (orange) of unlabeled, protonated Rai1. The resonances in the active site of Xrn2 are not influenced, indicating that Rai1 does not affect the active site of Xrn2. (C) Structure of Xrn2. Ileδ1 methyl groups that show pronounced changes in the HMQC spectrum upon interaction with Rai1 are highlighted in orange. The putative Rai1 interaction surface, inferred from the structure of the Xrn2:Rai1 complex in S. pombe (PDB 3FQD), is highlighted in blue. CSPs upon Rai1 interaction reach from the Rai1 binding site towards a region that is remote from the active site. pronounced changes in the HMQC spectrum upon interaction with Rai1 are highlighted in orange. The putative Rai1 interaction surface, inferred from the structure of the Xrn2:Rai1 complex in S. pombe (PDB 3FQD), is highlighted in blue. CSPs upon Rai1 interaction reach from the Rai1 binding site towards a region that is remote from the active site.