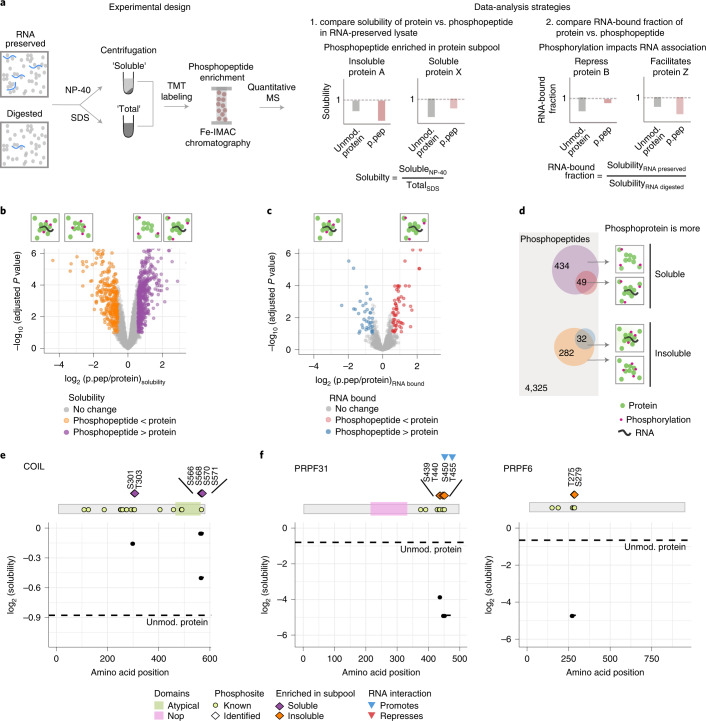
Fig. 2. Mapping phosphorylation sites of distinct protein pools.
a, Schematic representation of the experimental design and data-analysis strategies. Fe-IMAC, Fe3+-immobilized metal ion affinity chromatography; p.pep, phosphopeptide; TMT, tandem mass tag; unmod., unmodified. b, Volcano plot of the differential solubility of phosphopeptides of a protein compared to its unmodified protein in RNA-preserved lysate. Phosphopeptides exhibiting significantly (|log2 (FC)| > 0.5 and adjusted P value obtained from limma analysis (Benjamini–Hochberg) < 0.01) lower (orange) and higher (purple) solubility than the unmodified proteins are shown. c, Volcano plot of the differential RNA-bound fraction of phosphopeptides of a protein compared to its unmodified protein. Phosphopeptides exhibiting significantly (|log2(FC)| > 0.5 and adjusted P value obtained from limma analysis (Benjamini–Hochberg) < 0.01) lower (red) and higher (blue) proportions in the RNA-bound subpool than the unmodified proteins are shown. d, Venn diagram summarizing the overlap in different categories of assigned phosphopeptides. e,f, Visualization of the median solubility profiles (n = 3) of identified phosphopeptides (solid lines, phosphosites as points) and unmodified protein (dashed line) in log2 scale is represented along the linear sequence of the protein (x axis) of COIL (e), PRPF6 and PRPF31 (f). Top, schematic representation of the protein with its domains and known phosphosites from UniProt.