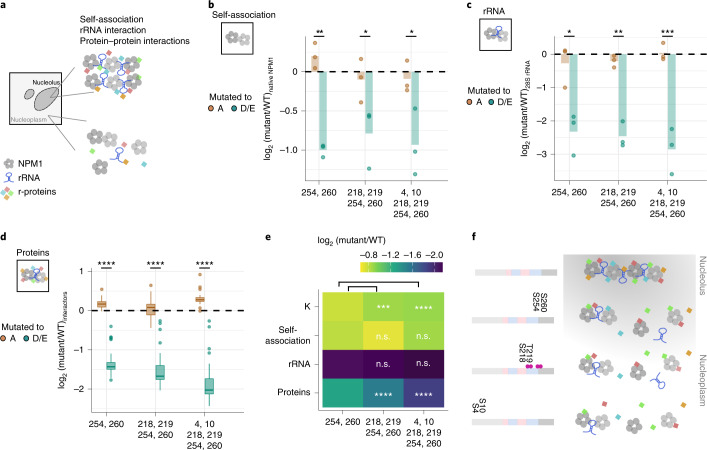
Fig. 6. Phosphorylation impairs both homotypic and heterotypic interactions of NPM1.
a, Schematic representation of the molecular interactions of NPM1 within the nucleolus and the nucleoplasm. b, Comparison of the self-association property (in log2 scale, y axis) of the indicated SNAP-tagged phosphomutants of NPM1 (x axis) normalized to that of SNAP-tagged NPM1 WT. This was measured by assessing the amount of HeLa cell (native) NPM1 associated with heterologously overexpressed SNAP-tagged NPM1 variants following immunoprecipitation (IP) with SNAP-tag as bait. Points represent data from three independent trials and the bar represents the mean value. Significance for each phosphomutant pair was calculated using Student’s t-test (two sided) and is represented by *P < 0.05, **P < 0.01 and ***P < 0.001. c, Comparison of the amount of 28S rRNA (in log2 scale, y axis) associated with the indicated SNAP-tagged phosphomutants of NPM1 (x axis) normalized to that of SNAP-tagged NPM1 WT following IP with SNAP-tag as bait. Points represent data from three independent trial and the bar represents the mean values. Significance for each phosphomutant pair was calculated using Student’s t-test (two sided) and is represented by *P < 0.05, **P < 0.01 and ***P < 0.001. d, Comparison of the relative amounts of NPM1-interacting proteins (in log2 scale, y axis; 44 proteins were classified as NPM1 interactors, including both ribosomal and non-ribosomal proteins) associated with the indicated SNAP-tagged phosphomutants of NPM1 (x axis) in comparison to those associated with SNAP-tagged NPM1 WT following IP with SNAP-tag as bait. Significance for each phosphomutant pair was calculated using Student’s t-test and is represented by *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001. The box plots display the median and the IQR, with the upper whiskers extending to the largest value ≤1.5 × IQR from the 75th percentile and the lower whiskers extending to the smallest values ≤1.5 × IQR from the 25th percentile. e, Heatmap showing normalized relative effect sizes of partition coefficient values (K) and amount of native NPM1 (reflects NPM1 self-association), rRNA and protein interactors of NPM1 associated with the indicated phosphomimetic mutants (x axis). Significance levels were obtained with Student’s t-test (two sided) and are represented by *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001. f, Schematic representation of the working model of the impact of phosphorylation on NPM1’s propensity to form a biomolecular condensate. Pink dots represent phosphosites, gray circles indicate NPM1, diamond shapes represent r-proteins, and the stem-loop schematic represents rRNA.