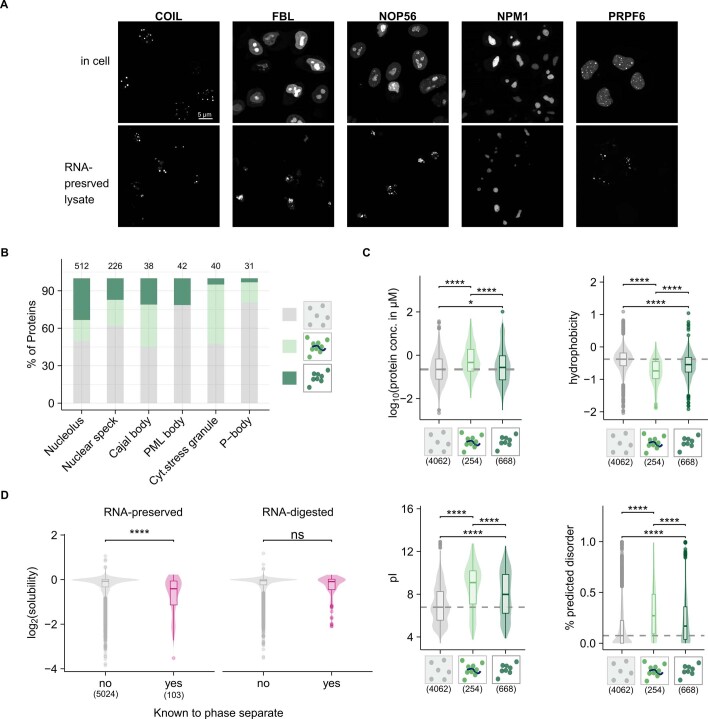
Extended Data Fig. 2. Physiochemical properties of proteins in different solubility subgroups.
(a) Representative confocal images of intact HeLa cells expressing GFP-tagged COIL, FBL, NOP56, NPM1 and PRPF6 and post-lysis using conditions used for proteomics assay. (b) Bar plot representing the proportion of proteins of different solubility classes present among proteins that are annotated to be part of various membrane-less organelles. Gene ontology annotation only based on experimental evidence was used for binning the proteins in different cellular compartments. Number of proteins from each organelle is shown on the top. (c) Distribution of intracellular protein concentration (top left, in log10 scale), hydrophobicity (top right, Kyte Doolittle scale), isoelectric point (pI, bottom left) and %predicted disorder in the sequence (bottom right) of proteins that were classified as ‘predominantly soluble’, and has an insoluble sub-pool that is ‘RNase-sensitive’ or ‘RNase-insensitive’. The box plots display the median and IQR, with the upper whiskers extending to the largest value ≤1.5 × IQR from 75th percentile and the lower whiskers extending to smallest values ≤1.5 × IQR from 25th percentile. Numbers represent the number of proteins in each category. Significance calculated using Wilcoxon signed-rank test (two-sided) and represented as ns: not significant, *p < 0.05, **p < 0.01, and ***p < 0.001. (d) Distribution of solubility (in log2 scale) of proteins that are known to undergo phase separation based on in-vitro experiments (curated list from PhaseDB) in RNA-preserved (left) and RNA-digested (right) lysate. Numbers represent the number of proteins in each category. Significance calculated using Wilcoxon signed-rank test (two-sided) and represented as ns: not significant, *p < 0.05, **p < 0.01, and ***p < 0.001.