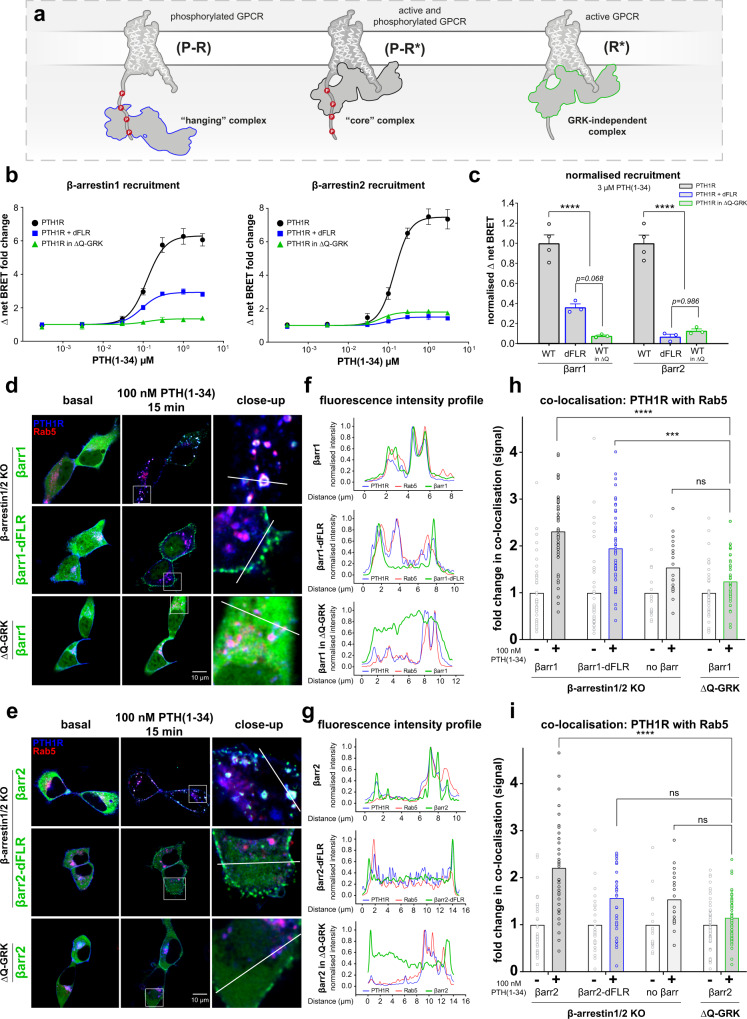
Fig. 1. The configuration of specific PTH1R–β-arrestin complexes determines their functionality.
a Schematic depiction of the ‘hanging’, ‘core’ and GRK-independent GPCR-β-arrestin complex configurations. b NanoBRET-measured recruitment of β-arrestin1 and 2 to the PTH1R upon stimulation with indicated concentrations of PTH(1-34). Curves show the recruitment measured for β-arrestin WT constructs in HEK293 cells (HEK-WT), analogous experiments performed in ΔQ-GRK cells and recruitment measurements of β-arrestin-dFLR mutants in HEK-WT. Results are shown as Δ net BRET fold change, mean of at least three independent repetitions (βarr-dFLR and βarr in ΔQ-GRK n = 3; βarr-WT n = 4) ± SEM. c Data correspond to the BRET changes at saturating ligand concentration from b, normalised to the respective β-arrestin WT condition. To test for significance between β-arrestin association in a ‘hanging’ or GRK-independent complex, a one-way ANOVA, followed by a two-sided Tukey’s test (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001). For both β-arrestin 1 and 2, WT vs respective βarr-dFLR and βarr in ΔQ-GRK p < 0.0001. d and e Representative live-cell confocal microscopy images of β-arrestin1/2 double knockout and ΔQ-GRK cells transfected with PTH1R-CFP (blue), the early endosome marker Rab5-mCherry (red) and the respective β-arrestin-YFP WT or dFLR constructs (green). Additionally, close-ups of the stimulated conditions are shown, which correspond to the white squares indicated in the representative image. Images were acquired before and after stimulation with 100 nM PTH(1-34) for 15 min from at least three cover slips, prepared from at least three independent transfections (n ≥ 3). f and g normalised fluorescence intensity profiles of all three acquired channels along the white line indicated in the close-up images in d or e, respectively. h and i The quantification of co-localisation of PTH1R-CFP with Rab5-mCherry in β-arrestin1/2 knockout and ΔQ-GRK cells was calculated using Squassh and SquasshAnalyst (number of images per respective condition; βarr1 (39), βarr1-dFLR (43), no βarr (17), βarr1 in ΔQ-GRK (38), βarr2 (33), βarr2-dFLR (27), βarr2 in ΔQ-GRK (50)) and is represented as mean fold change in co-localisation signal + SEM. Statistical significance was calculated by two-way ANOVA, followed by a post-hoc comparison with Bonferroni correction (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001).Complete results of the statistical analysis are shown in Supplementary Table 1. Source data are provided as a source data file.