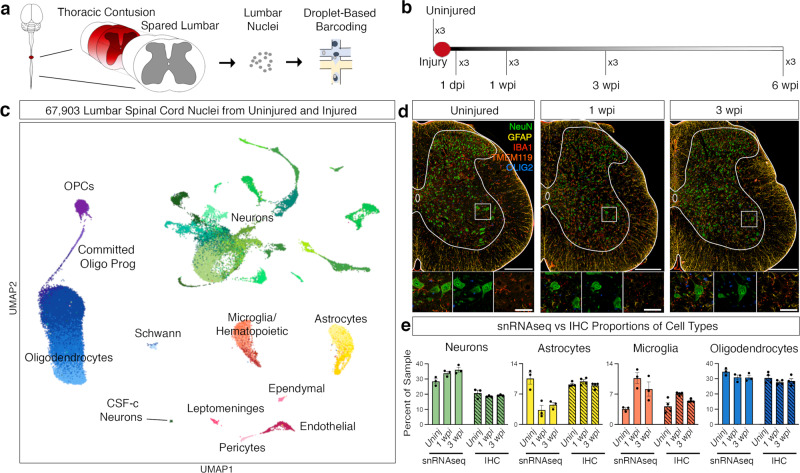
Fig. 1. Single nucleus RNA sequencing of the lumbar spinal cord after thoracic contusion.
a Schematics depicting the experimental design for snRNA-seq, showing the injured thoracic cord and lumbar cord (with dark red representing the lesion) as well as nuclei isolation from the intact lumbar cord followed by droplet-based barcoding for single nucleus RNA sequencing. b Top, an overview of experimental design for injury and tissue collection. The lumbar spinal cord of three animals from each time point: uninjured, 1 dpi (day post injury), 1 wpi (week post injury), 3 wpi, and 6 wpi. c Uniform manifold approximation and projection (UMAP) visualization of 67,903 nuclei from uninjured and injured lumbar spinal cords, revealing 9 classes and 39 subtypes. Colored by green (neurons), yellow, astrocytes, orange-red (microglia), purple (OPCs), blue (oligodendrocytes), light blue (Schwann), light pink (pericytes), pink (ependymal), magenta (leptomeninges), brick-red (endothelial). d Multiplex immunohistochemistry (IHC) of the lumbar spinal cord from uninjured, 1 wpi and 3 wpi. Tissue was stained for NeuN (green), GFAP (yellow), IBA1 (red), TMEM119 (dark orange), and OLIG2 (blue). Scale bars are 200 µm (main) and 50 µm (inset), respectively. e Quantification of the proportion of cell types from the snRNA-seq data and immunohistochemistry in tissue. Mean ± SEM; snRNAseq, N = 3; immunohistochemistry, N = 4. Source data are provided as a Source Data file.