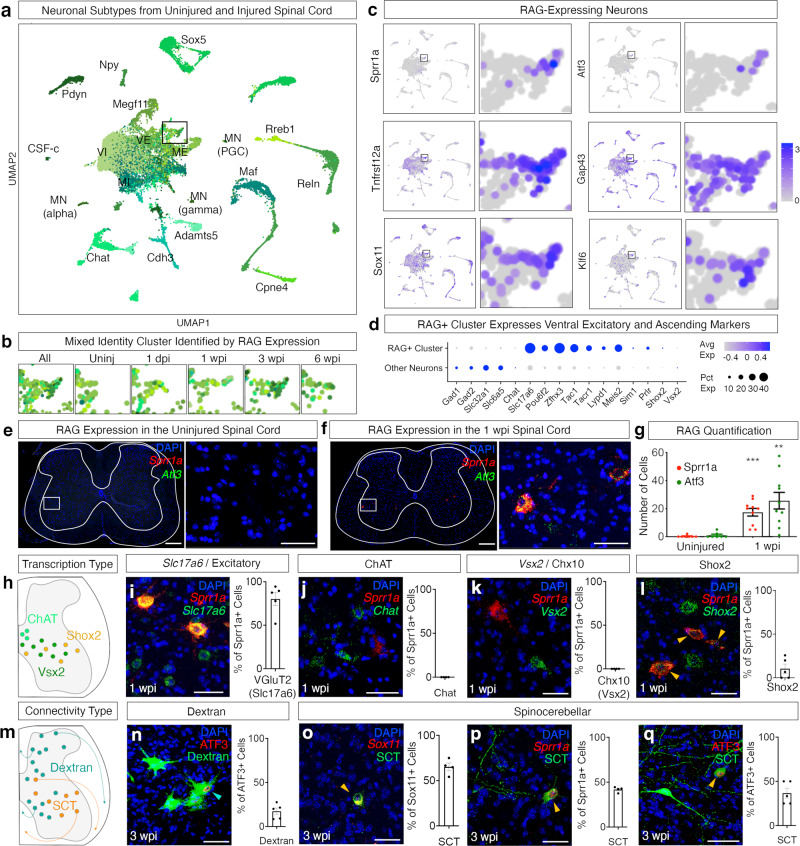
Fig. 4. Specific neurons express genes associated with regeneration.
a UMAP showing predicted neuronal families. b Targeted view of RAG-expressing cluster over injury time points. c Featureplots showing RAGs expressed in neurons. d A dotplot showing expression of RAGs within targeted RAG-expressing cluster (cluster 23 defined by independent clustering in Supplementary Fig. 5). Average expression (avg exp) is indicated by color (gray to blue) and percent expressed (pct exp) is indicated by the size of the circle. e, f RNAscope in situ hybridization showing expression of Sprr1a and Atf3 in the uninjured cord and 1 wpi. Scale bars are 200 and 50 µm, respectively. g Quantification of Sprr1a+ cells and Atf3+ cells in the uninjured and 1 wpi injury cord (p = 0.001 shown as ***p = 0.0037 shown as **, two-sided unpaired t-test, Error bars indicate ± SEM, N = 7, 10). h Diagram of spatial location of transcription types, including ChAT (light green), Vsx2 (dark green), and Shox2 (orange). i–l Visualization and quantification of VGluT2/Slc17a6+, Shox2+, Chx10/Vsx2+, and Sprr1a-expressing cells in the ventral spinal cord. Scale bars are 50 µm. Error bars indicate ± SEM (N = 5, 4, 4, 6 animals). m Diagram of spatial location of connectivity types, including ascending neurons labeled by dextran (aqua) and spinocerebellar (SCT, orange) neurons. n Visualization and quantification of lumbar spinal cord neurons labeled by dextran injected into a thoracic contusion lesion site. Aqua arrows indicate ATF3 and dextran overlap. Scale bars are 50 µm. Error bars indicate ± SEM (N = 5 animals). Visualization and quantification of the percent of RAG-expressing cells—Sox11 (o), Sprr1a (p), and ATF3 (q) that are spinocerebellar. Spinocerebellar neurons are shown in green and RAGs are shown in red. Orange arrows indicate RAG gene and spinocerebellar dual-labeled cells. Scale bars are 50 µm. Error bars indicate ± SEM (N = 4, 4, 5 animals). Source data are provided as a Source Data file.