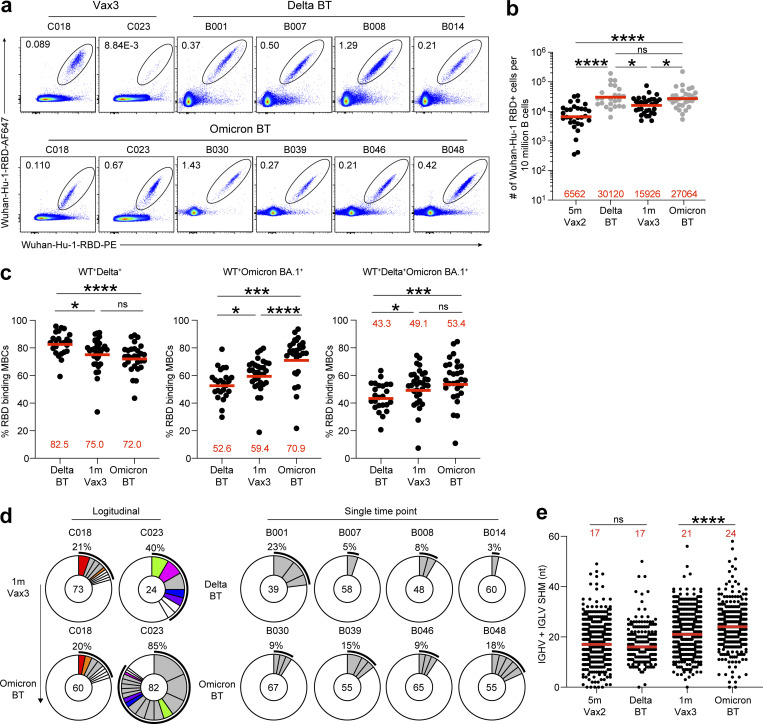
Figure 2.
Anti–SARS-CoV-2 RBD MBCs after breakthrough infection. (a) Representative flow cytometry plots indicating PE-WT-RBD and AlexaFluor-647–WT-RBD binding MBCs from four individuals after Delta breakthrough infection following Vax2 (Delta BT), two individuals 1 mo after Vax3, and six individuals after Omicron BA.1 breakthrough infection following Vax3 (Omicron BT). (b) The number of WT RBD–specific B cells is indicated, 5 mo (m) after Vax2 (Cho et al., 2021), Delta BT (n = 24), 1 mo after Vax3 (Muecksch et al., 2022), and Omicron BT (n = 29). (c) Graphs showing the percentage of WT-, Delta-, and Omicron BA.1-RBD cross-binding B cells determined by flow cytometer in vaccinees (Vax3) and breakthrough individuals (Delta BT or Omicron BT; see also Fig. S2 b). (d) Pie charts show the distribution of IgG antibody sequences obtained from WT-specific MBCs from: two individuals assayed sequentially 1 mo after the third mRNA dose (Vax3) and followed by an Omicron infection (Omicron-BT; left); four individuals after Delta breakthrough (Delta); and four individuals after Omicron breakthrough (Omicron). The number inside the circle indicates the number of sequences analyzed for the individual denoted above the circle. Pie slice size is proportional to the number of clonally related sequences. The black outline and associated numbers indicate the percentage of clonal sequences detected at each time point. Colored slices indicate persisting clones (same IGHV and IGLV genes, with highly similar CDR3s) found at more than one time point within the same individual. Gray slices indicate clones unique to the time point. White slices indicate sequences isolated only once per time point. (e) Number of nucleotide somatic hypermutations (SHM) in IGHV + IGLV in WT-RBD–specific sequences after Delta or Omicron breakthrough infection, compared to 5 mo after Vax2, and 1 mo after Vax3. All experiments were performed at least in duplicate and repeated twice. Red bars and numbers in panels b and c represent geometric mean, and in panel e represent median values. Statistic analysis in panels b and c was determined by two-tailed Kruskal–Wallis test with subsequent Dunn’s multiple-comparisons test and in panel e by two-tailed Mann–Whitney test. *, P ≤ 0.05; ***, P ≤ 0.001; ****, P ≤ 0.0001; ns, not significant.