Figure 1. A high‐content screen maps the peroxi‐ome.
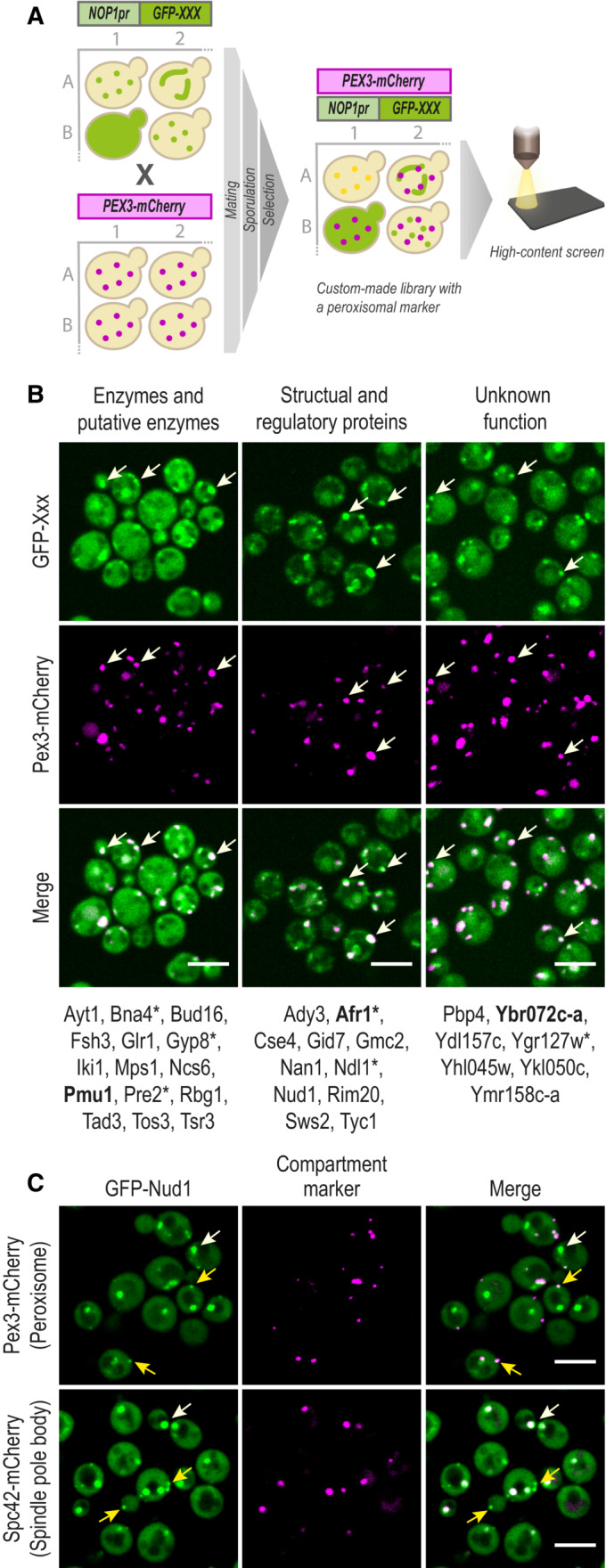
- A peroxisomal marker Pex3‐mCherry was genomically integrated into a yeast N′ GFP collection by utilizing an automated mating procedure. Then, high‐throughput fluorescence microscopy was applied to identify proteins that co‐localize with the peroxisomal marker. Only proteins that passed three validation steps (manual retagging, PCR, and western blot (Appendix Fig S1)) were determined as newly identified peroxisomal proteins.
- The validated proteins were divided into groups to represent their variety of functional annotations: enzymatic, structural, or unknown activity. In bold are the proteins whose images are presented. All proteins were tested for their localization when their encoding genes are expressed under their native promoter (Dataset EV1); asterisks mark the ones that could be visualized in peroxisomes.
- Protein localization image analysis demonstrated that about half of the newly identified proteins are dually localized to peroxisomes and other compartments. Presented is GFP‐Nud1, which co‐localizes with both a spindle‐pole body marker (white arrows) and a peroxisomal marker (yellow arrows). A full analysis of dual‐localization is shown in Appendix Fig S2 and Dataset EV1.
Data information: For all micrographs, a single focal plane is shown. The scale bar is 5 μm.
