Fig. 1. Activity of SIV Env-specific mAbs.
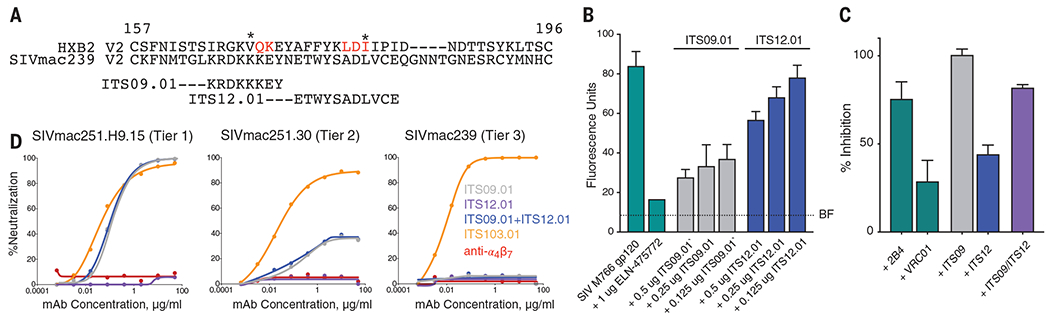
(A) Alignment of the amino acid sequences of the V2 domains of HIV HXB2 and SIVmac239. The regions of SIV V2 containing epitopes recognized by the SIV V2 mAbs ITS09.01 and ITS12.01 are indicated. Amino acids identified as part of the α4β7 binding site in HIV V2 are indicated in red. Asterisks denote the positions of V2 sieve residues in the RV144 vaccine trial. (B) Adhesion of α4β7-expressing RPMI8866 cells to SIVmac766 gp120 in the presence of decreasing concentrations of ITS09.01 and ITS12.01. Adhesion of cells to gp120 alone or in the presence of the α4β7 antagonist ELN-475722 is included. BF indicates background fluorescence in the absence of cells. Conditions are carried out in triplicate; error bars indicate SD. (C) Inhibition of α4β7 adhesion by the combination of IT12.01 and ITS09.01. The α4β7 mAb 2B4 is used as a positive specificity control (14), and VRC01, an antibody to HIV that does not bind SIV, is used as a negative nonspecific reagent control. For (B) and (C), results shown are representative of three independent experiments and indicate percentage inhibition relative to adhesion in the absence of any mAb. Conditions are run in triplicate and error bars indicate ±1 SD. (D) ITS09.01, ITS12.01, and anti-α4β7 do not neutralize the Tier 3 SIV mac239. ITS09.01 shows partial neutralization of the Tier 2 virus, and complete neutralization of the Tier 1 clone. By contrast, ITS103.01, which targets the CD4 binding site, completely neutralizes nearly all strains of SIV including SIVmac239. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
