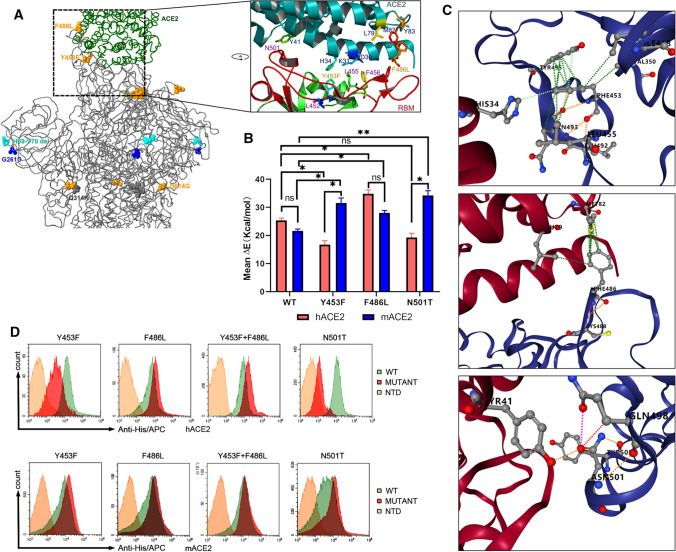
Fig. 3.
Analysis of binding of the SARS-CoV-2 spike protein with human and mink receptors. (A) Comparison of the spike structure of mink SARS-CoV-2 with that of reference strain WIV04. The changed residues within mink SARS-CoV-2 are highlighted as yellow balls. (B) The free energy of binding of the wild-type RBD or mutants from mink SARS-CoV-2 to the human and mink receptor. (C) Amino acid changes involved in the stability of the RBD-ACE2 complex. Detailed structures for Y453, F486, and N501 are arranged from top to bottom. The green lines represent hydrophobic interactions, the orange lines indicate polar H-bonds, the red lines represent hydrogen bonds, and the pink-purple lines represent clashes. (D) Measurement of the binding of RBD mutants to human ACE2 (hACE2, upper panel) and mink ACE2 (mACE2, lower panel) by FACS. His-tagged wild-type RBD, RBD mutants, and NTD were incubated with cells expressing eGFP-fused ACE2. NTD was used as a negative control.