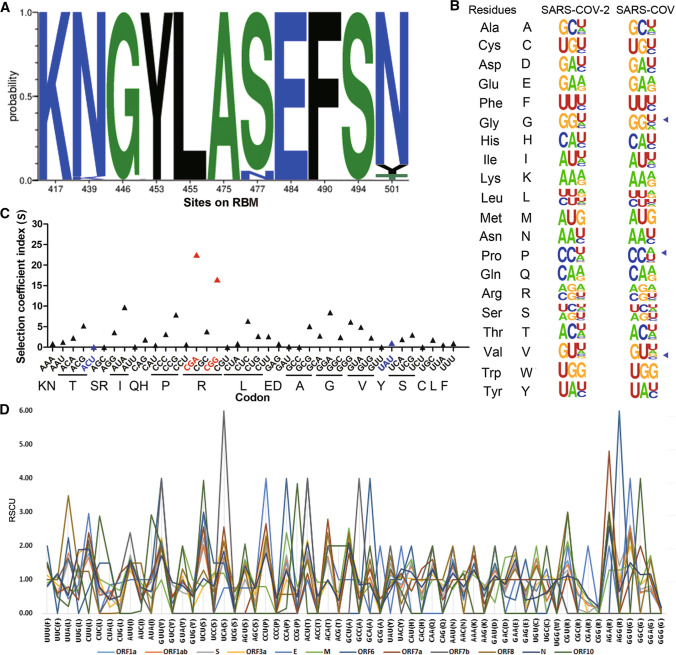
Fig. 4.
Codon usage and fitness analysis of the SARS-CoV-2-encoded proteins. (A) Probability of mutations in the receptor-binding motif (RBM). The data were analyzed using MEGA X software. The frequency was calculated using the Datamonkey server, and the figure was produced using WebLogo (https://weblogo.berkeley.edu/logo.cgi). (B) The synonymous codon usage of SARS-CoV-2. The figure was produced using WebLogo. The mink SARS-CoV-2 sequence (GenBank no. MT396266) was compared with that of SARS-CoV strain Tor2 (GenBank no. NC_004718.3). (C) Selective coefficient index for SARS-CoV-2 codons. The codons for the N501 mutants are shown in blue and the codons with the highest fitness are highlighted in red. (D) Analysis of relative synonymous codon usage in SARS-CoV-2-encoded proteins.