FIGURE 1.
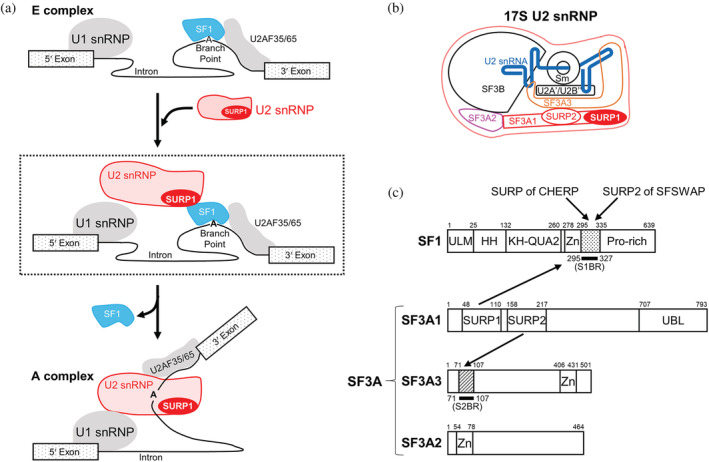
Splicing factors involved in early spliceosome assembly. (a) Schematic diagram of the process of transition from the E to A complex. U2 snRNP binds to SF1 via SURP1 for the recruitment of U2 snRNP to the E complex, shown in the dashed box. During the E to A conversion process, which occurs in an ATP‐dependent manner, SF1 is displaced by U2 snRNP and released from the spliceosome. (b) Schematic diagram of 17S U2 snRNP. U2 snRNA is represented by a blue bold line. SF3A1, SF3A2, and SF3A3 proteins, which are components of the SF3A protein complex, are depicted in red, magenta, and orange, respectively. Two tandem SURP domains, SURP1, and SURP2, in SF3A1 are represented by red oval shapes. (c) Schematic diagrams of the domain architecture of SF1, SF3A1, SF3A2, and SF3A3 from humans. SF1 has five structural domains, namely, the UHM ligand motif (ULM), helix hairpin (HH), K homology and Quaking homology 2 domain (KH‐QUA2), and zinc‐finger domain (Zn). In addition, SF1 has a proline‐rich region (Pro‐rich) in the C terminus. Regarding the SF3A complex, SF3A1 has three structural domains, SURP1, SURP2, and a Ubiquitin‐like domain (UBL) whilst SF3A2 and SF3A3 have a single structural domain, a zinc‐finger domain (Zn). No other known motifs have been found in the three components of SF3A. Bold black lines indicate a “SURP1‐binding site”‐containing region (S1BR, dotted) and the SURP2 counterpart (S2BR, hatted). A peptide corresponding to each region was used to determine the structures of the SURP1 and SURP2 complexes. Each of the binding partners of the four SURP domains is indicated by an arrow; SURP1 of SF3A1 and the SURP domains of CHERP and SFSWAP bind to S1BR in SF1, while SURP2 binds to S2BR in SF3A3.
