FIGURE 2.
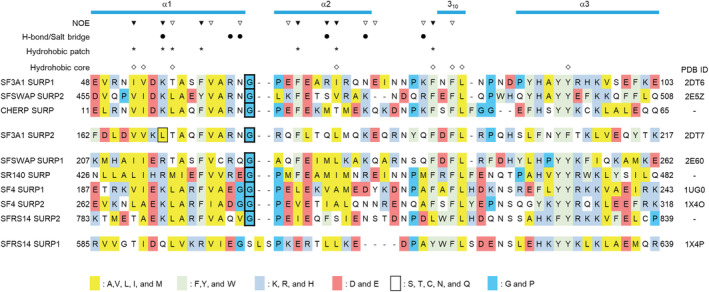
Structure‐based sequence alignment of SURP domains from humans. Amino acid sequence alignments of SURP domains from humans were performed using the ClustalW program 55 and then manually modified based on the structures of the SURP domains. The accession codes used in the sequence alignment were as follows: SF3A1 (UniProt accession No. Q15459), SFSWAP (Q12872), CHERP (Q8IWX8), SR140 (O15042), SF4 (Q8IWZ8), and SFRS14 (Q8IX01). All structures were previously published by our group, and each protein data bank (PDB) code is shown. The secondary structure elements are shown in cyan at the top. The alignments are colored according to the amino acid type shown in the alignment legend. Each box with thick black lines indicates a Gly residue connecting α1 and α2, which forms the G bend. The single box with thin black lines indicates Leu169 in α1 of SURP2, which is the determinant of the SURP2 complex formation. The inverted triangles indicate SURP1 residues for which intermolecular NOEs from S1BR were observed (open inverted triangle: less than 10 NOEs, filled inverted triangle: 10 or more NOEs). Filled round marks indicate SURP1 residues that participate in hydrogen bonds or salt bridges with S1BR. Asterisks indicate SURP1 residues that form a hydrophobic patch that interacts with S1BR. Diamonds indicate residues that form a hydrophobic core common to SURP domains.
