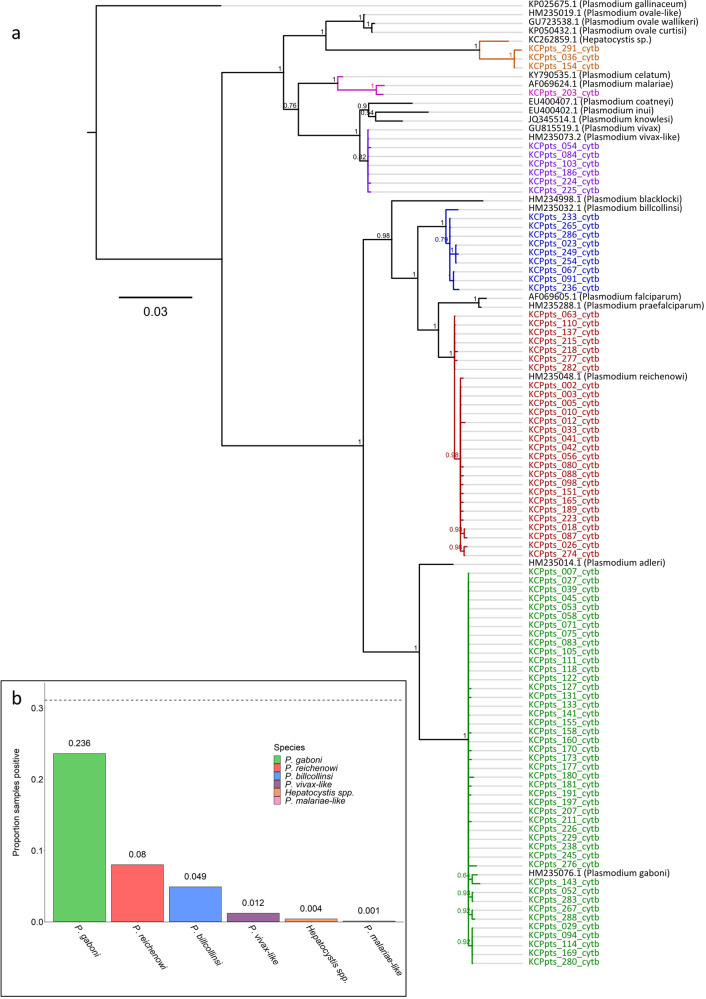
Fig. 2. Richness of malaria parasites isolated from the Kanyawara chimpanzee community.
a Bayesian phylogeny generated from a representative subset of the SGA-derived cytB sequences generated in this study. Sequences were derived from N = 878 fecal samples, collected from the Kanyawara chimpanzee community in Kibale National Park, western Uganda, between 2013 and 2016. Color corresponds to malaria parasite species, inferred from the phylogenetic relationships of sequences to previously published reference sequences (green: P. gaboni, red: P. reichenowi, blue: P. billcollinsi, violet: P. vivax-like, orange: Hepatocystis spp., magenta: P. malariae). Newly generated sequences are listed in Supplementary Data 3, and previously published reference sequences used in this figure are listed in Supplementary Data 1. b Distribution of parasite species isolated from the Kanyawara chimpanzees. Malaria parasites were amplified in 31.1% of samples (dotted line). A majority of parasites amplified were members of the sub-genus Laverania (i.e., relatives of P. falciparum): P. gaboni (23.6%), P. reichenowi (8.0%), and P. billcollinsi (4.9%). Members of the primate malaria clade were amplified in a minority of samples: P. vivax-like (1.2%) and P. malariae-like (0.1%). In addition, Hepatocystis was amplified from 0.4% of samples.