Figure 4.
The N460K mutation determines enhanced cell-cell fusion and S processing of BA.2.75
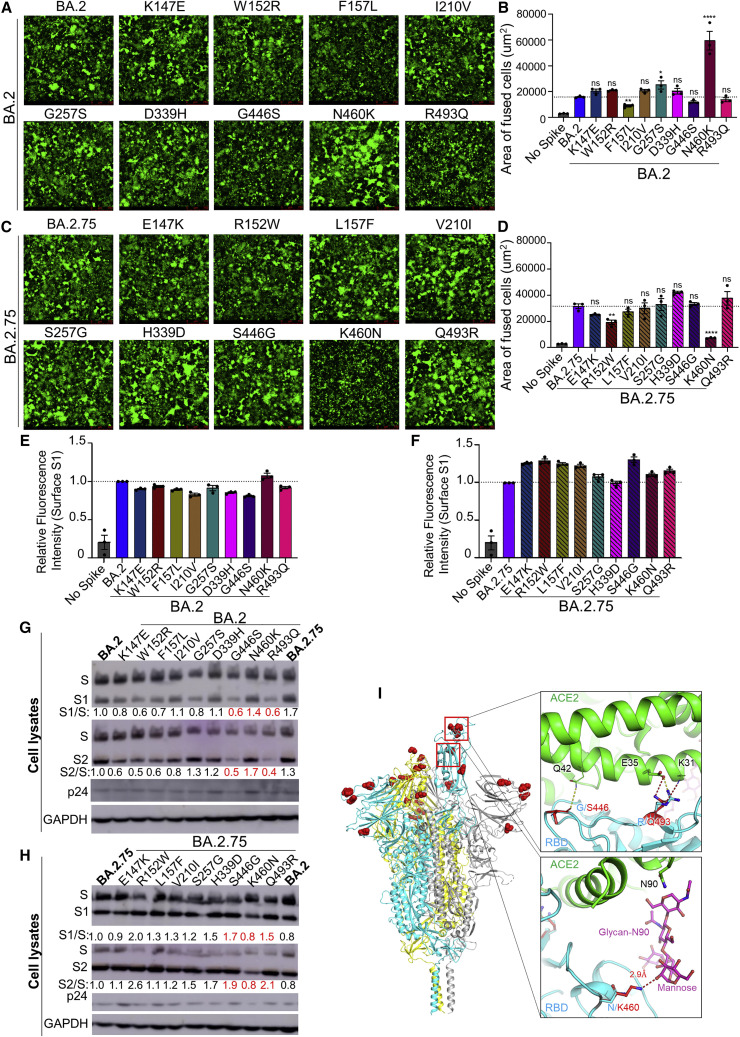
(A) Fluorescence images displaying syncytia formation are presented for HEK293T-ACE2 cells 48 h after co-transfection with a GFP expression construct and BA.2 single mutant S proteins. Scale bars represent 150 μm.
(B) Quantification of syncytia formation in (A) displays the mean syncytia size; bars represent mean ± standard error, with significance relative to D614G determined by one-way ANOVA with Bonferroni multiplicity correction with p values displayed as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, and ns for not significant.
(C) Fluorescence images displaying syncytia formation are presented for HEK293T-ACE2 cells 48 h after co-transfection with a GFP expression construct and BA.2.75 single reversion mutant S proteins. Note that the negative control for Figures 4A and 4C corresponds to that shown in Figure 3A, which were from the same experiment. Scale bars represent 150 μm.
(D) Quantification of syncytia formation in (C) displays the mean syncytia size; bars represent mean ± standard error, with significance relative to D614G determined by one-way ANOVA with Bonferroni multiplicity correction with p values displayed as ∗∗p < 0.01, ∗∗∗∗p < 0.0001, and ns for not significant.
(E and F) Quantification of relative S surface expression in transfected HEK293T cells for BA.2 single mutants (E) or BA.2.75 reversion mutants (F), as examined by flow cytometry; bars represent mean ±standard error.
(G) Pseudotyped lentivirus producer cell lysate was assessed for processing of S from BA.2 single mutants by probing with anti-S1 (T62), anti-S2, anti-HIV-1 p24, and anti-GAPDH. Band intensities were quantified in ImageJ and the ratios of S1/S and S2/S are displayed relative to the S1/S and S2/S ratios of BA.2.
(H) Pseudotyped lentivirus producer cell lysate was assessed for processing of S from BA.2.75 reversion mutants by probing with anti-S1, anti-S2, anti-HIV-1 p24, and anti-GAPDH. Band intensities were quantified in ImageJ and the ratios of S1/S and S2/S are displayed relative to the S1/S and S2/S ratios of BA.2.75.
(I) Structural modeling of Omicron BA.2.75 spike protein viewed as a ribbon. Mutations of BA.2.75 specific mutants are highlighted by red spheres. The RBD of the cyan spike protomer is in an “up” conformation. Upper inset: the mutation G446S reduces the backbone flexibility and possibly stabilizes the hydrogen bond between its carbonyl group and the residue Q42 on ACE2 receptor (green); the mutation R493Q abolishes the salt bridge interaction with the E35 on ACE2 receptor and potentially forms two hydrogen bonds with E35 and K31. Lower inset: the mutation N460K enables formation of a hydrogen bond with the glycan-N90 on ACE2 receptor (green).