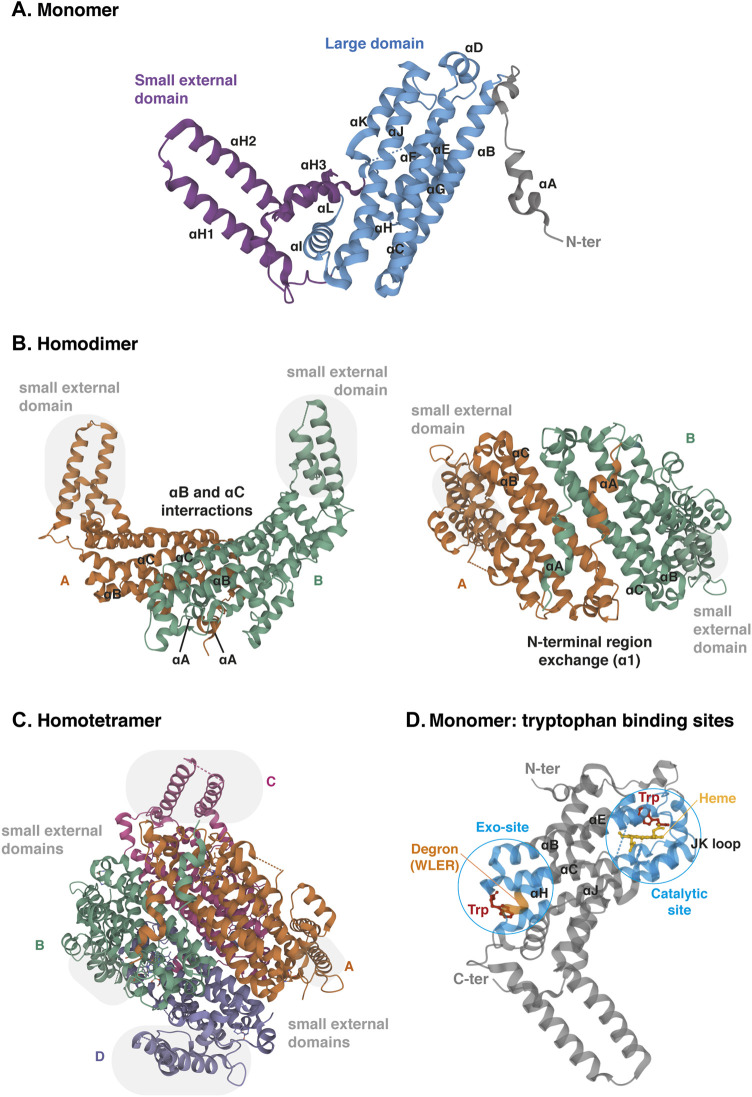
FIGURE 5.
Crystal structure of human TDO (apo protein). (A) Monomeric conformation of TDO illustrating the secondary structure comprising fifteen α-helices distributed in three parts: the large central domain (blue), the small external domain (purple) and the N-terminal region (grey). α-Helices are labeled αA- αL according to the TDO structure described by Lewis-Ballester and colleagues (Lewis-Ballester et al., 2016). (B) Homodimeric conformation of TDO in two orientations. Monomers are distinguished by two colors. TDO monomers are associated in pairs through the interactions of their α-helices B and C of the large domains and through the exchange of their α-helices A of the N-terminal regions. Grey highlight shows the small external domains not involved in the interaction of monomers A and B. (C) Homotetrameric conformation of TDO. Monomers are distinguished by four colors. Dimers are clamped together through the interaction of their α-helices J. Dimer interaction is stabilized by the exchange of the small external domains (grey highlight) grafted on the opposite large domains (A with C and B with D, and vice versa). (D) Monomeric conformation of TDO illustrating the two tryptophan-binding sites. The TDO catalytic site (blue, right) is surrounded by the bundle from helices αB, αC, αJ, αE and αH. The heme cofactor (yellow) is bound by the proximal His328 from αJ. The TDO exo-site (blue, left) is also surrounded by the four-helical bundle from αB, αC, αJ, αE and αH, opposite the catalytic site. The orange color inset indicates the four-amino acid degron masked by tryptophan, triggering TDO degradation in low tryptophanemia (Klaessens et al., 2021). The 3D structure of TDO was modelized with Mol* Viewer from the crystal structure of TDO described by Lewis-Ballester et al. (2016) (PDB file ID: 5TIA).