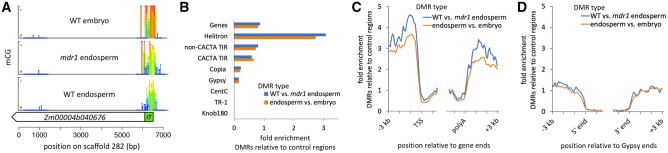
Figure 3.
A, Differential methylation of an r1 gene in embryo, endosperm, and mdr1 mutant endosperm. mCG values are shown at single-base level resolution over an 8-kb region including the majority of an r1 homolog in W22 (Zm00004b040676). Both height and color of lollipops indicate mCG values (darkest blue = zero mCG). Regions without lollipops indicate lack of read coverage, either because of multi-copy sequence or large N gaps. B, DMR enrichment in different genetic elements. Enrichment is the proportion of DMRs that overlap each element by at least 50% of their length (100 bp) divided by the proportion of control regions that do. Control regions are the 200-bp regions that were eligible for identifying DMRs based on read coverage and number of informative cytosines. C and D, Enrichment for DMRs on 100-bp intervals relative to gene ends (C) or Gypsy ends (D). Gypsies with solo LTRs were excluded from this analysis. For genes, TSS = Transcription start site, polyA = polyadenylation site.