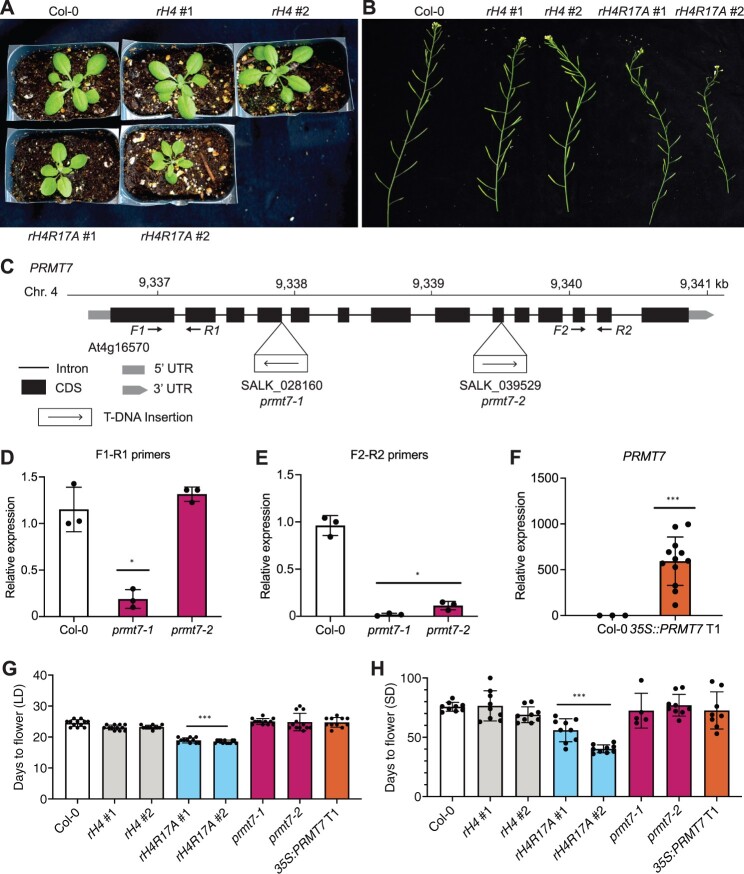
Figure 3.
PRMT7 does not regulate the floral transition in Arabidopsis. A, Rosette phenotype of Col-0, rH4 #1, rH4 #2, rH4R17A #1, and rH4R17A #2 plants grown in long-day conditions for 3 weeks. B, Silique phenotype of Col-0, rH4 #1, rH4 #2, rH4R17A #1, and rH4R17A #2 plants grown in long-day conditions for 4 weeks. C, Schematic diagram of the PRMT7 locus. The location of the T-DNA insertions and the primers (F1–R1 and F2–R2) used for gene expression analyses are shown. D–F, RT-qPCR showing relative PRMT7 transcript levels in Col-0, prmt7-1, and prmt7-2 plants (D and E), and Col-0 and 35S:PRMT7 T1 plants (F). The average of three biological replicates and standard deviation are shown for Col-0 and prmt7 mutants. For the 35S:PRMT7 plants, individual data points represent independent T1 plants. *P < 0.05; **P < 0.005; ***P < 0.0005, as determined by unpaired Student’s t test (sample versus Col-0). E and F, Mean days to flower in long-day (E) or short-day conditions (F) for Col-0, rH4 #1, rH4 #2, rH4R17A #1 and rH4R17A #2, prmt7-1, prmt7-2, and 35S:PRMT7 T1 plants. Error bars show standard deviation. *P < 0.05; **P < 0.005; ***P < 0.0005, as determined by one-way ANOVA with Tukey’s honestly significant difference (HSD) post hoc test. n ≥ 11 for long days, n ≥ 5 for short days.